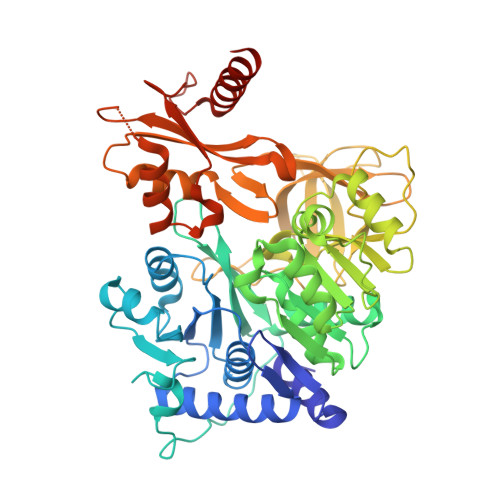
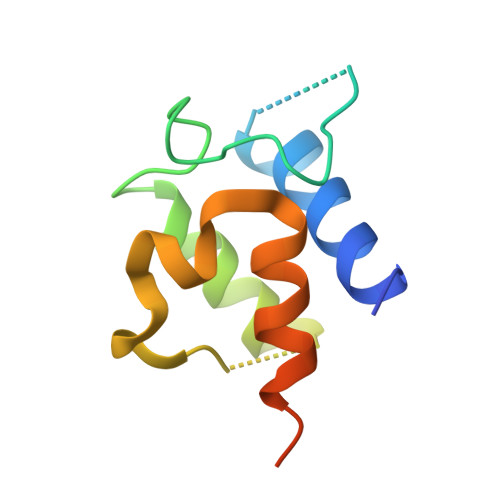
Dynamic visualization of type II peptidyl carrier protein recognition in pyoluteorin biosynthesis.
Corpuz, J.C., Podust, L.M., Davis, T.D., Jaremko, M.J., Burkart, M.D.(2020) RSC Chem Biol 1: 8-12
- PubMed: 33305272
- DOI: https://doi.org/10.1039/c9cb00015a
- Primary Citation of Related Structures:
6O6E - PubMed Abstract:
Using a covalent chemical probe and X-ray crystallography coupled to nuclear magnetic resonance data, we elucidated the dynamic molecular basis of protein recognition between the carrier protein and adenylation domain in pyoluteorin biosynthesis. These findings reveal a unique binding mode, which contrasts previously solved carrier protein and partner protein interfaces.
Organizational Affiliation:
Department of Chemistry and Biochemistry, University of California-San Diego, 9500 Gilman Drive, La Jolla, CA 92093-0358, USA.