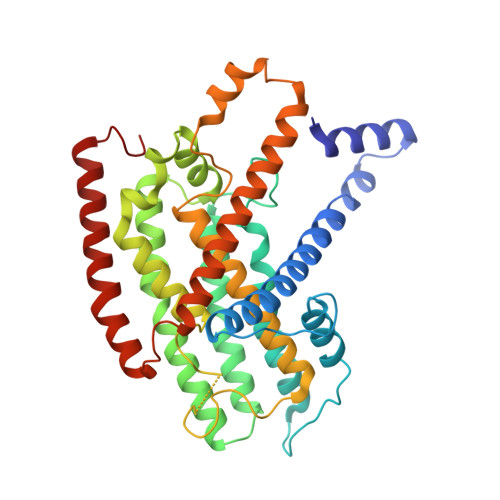
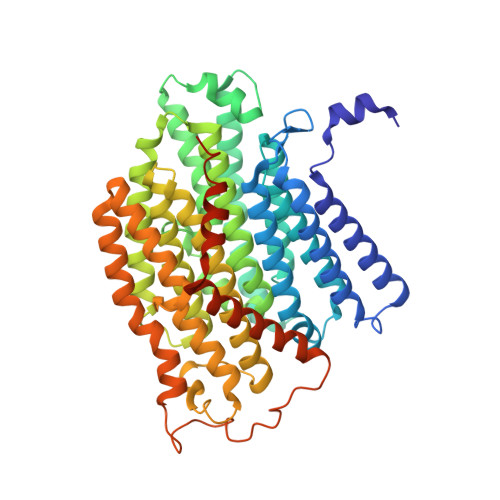
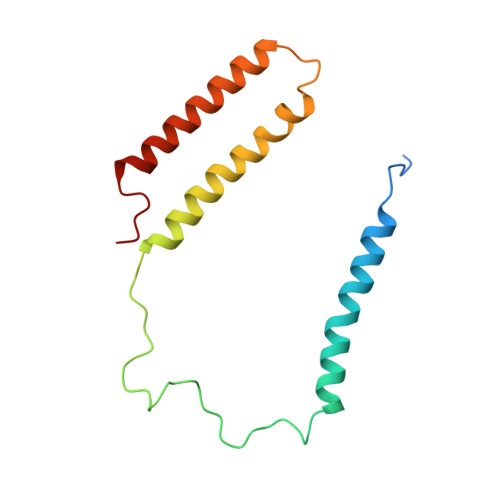
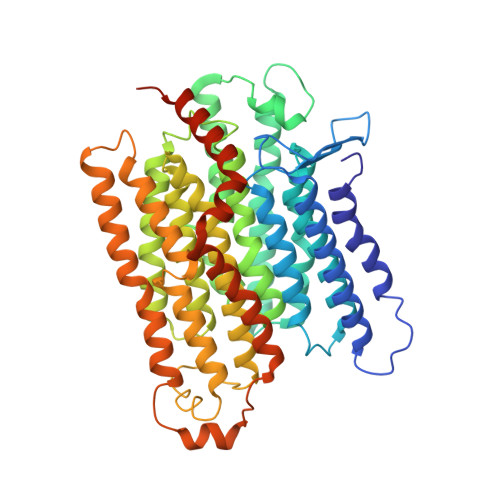
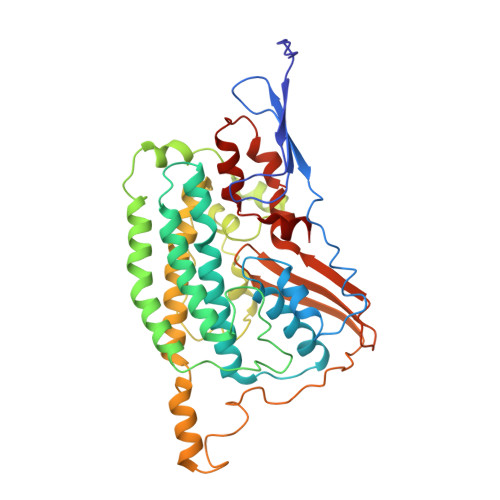
Structural insights into NDH-1 mediated cyclic electron transfer.
Zhang, C., Shuai, J., Ran, Z., Zhao, J., Wu, Z., Liao, R., Wu, J., Ma, W., Lei, M.(2020) Nat Commun 11: 888-888
- PubMed: 32060291
- DOI: https://doi.org/10.1038/s41467-020-14732-z
- Primary Citation of Related Structures:
6L7O, 6L7P - PubMed Abstract:
NDH-1 is a key component of the cyclic-electron-transfer around photosystem I (PSI CET) pathway, an important antioxidant mechanism for efficient photosynthesis. Here, we report a 3.2-Å-resolution cryo-EM structure of the ferredoxin (Fd)-NDH-1L complex from the cyanobacterium Thermosynechococcus elongatus. The structure reveals three β-carotene and fifteen lipid molecules in the membrane arm of NDH-1L. Regulatory oxygenic photosynthesis-specific (OPS) subunits NdhV, NdhS and NdhO are close to the Fd-binding site whilst NdhL is adjacent to the plastoquinone (PQ) cavity, and they play different roles in PSI CET under high-light stress. NdhV assists in the binding of Fd to NDH-1L and accelerates PSI CET in response to short-term high-light exposure. In contrast, prolonged high-light irradiation switches on the expression and assembly of the NDH-1MS complex, which likely contains no NdhO to further accelerate PSI CET and reduce ROS production. We propose that this hierarchical mechanism is necessary for the survival of cyanobacteria in an aerobic environment.
- Ninth People's Hospital, Shanghai Jiao Tong University School of Medicine, 200125, Shanghai, China.
Organizational Affiliation: