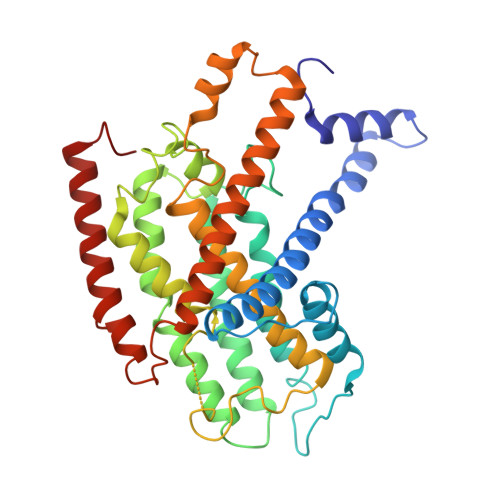
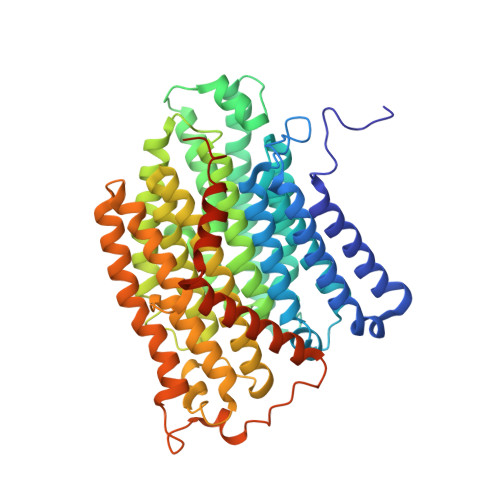
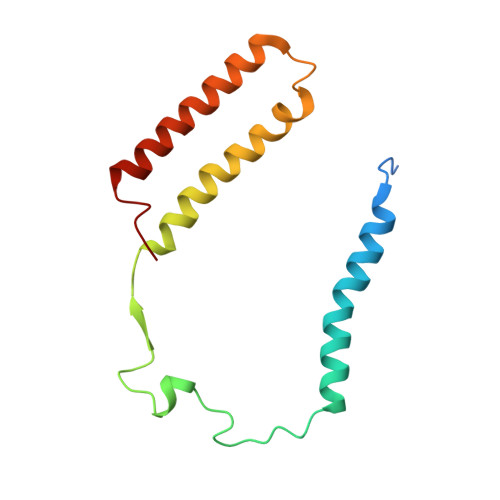
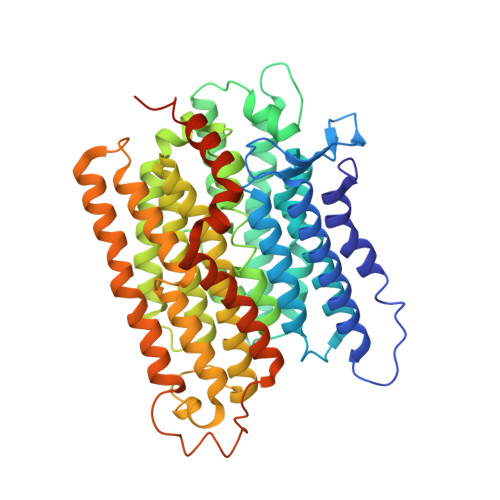
Structural basis for electron transport mechanism of complex I-like photosynthetic NAD(P)H dehydrogenase.
Pan, X., Cao, D., Xie, F., Xu, F., Su, X., Mi, H., Zhang, X., Li, M.(2020) Nat Commun 11: 610-610
- PubMed: 32001694
- DOI: https://doi.org/10.1038/s41467-020-14456-0
- Primary Citation of Related Structures:
6KHI, 6KHJ - PubMed Abstract:
NAD(P)H dehydrogenase-like (NDH) complex NDH-1L of cyanobacteria plays a crucial role in cyclic electron flow (CEF) around photosystem I and respiration processes. NDH-1L couples the electron transport from ferredoxin (Fd) to plastoquinone (PQ) and proton pumping from cytoplasm to the lumen that drives the ATP production. NDH-1L-dependent CEF increases the ATP/NADPH ratio, and is therefore pivotal for oxygenic phototrophs to function under stress. Here we report two structures of NDH-1L from Thermosynechococcus elongatus BP-1, in complex with one Fd and an endogenous PQ, respectively. Our structures represent the complete model of cyanobacterial NDH-1L, revealing the binding manner of NDH-1L with Fd and PQ, as well as the structural elements crucial for proper functioning of the NDH-1L complex. Together, our data provides deep insights into the electron transport from Fd to PQ, and its coupling with proton translocation in NDH-1L.
Organizational Affiliation:
National Laboratory of Biomacromolecules, CAS Center for Excellence in Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing, 100101, PR China.