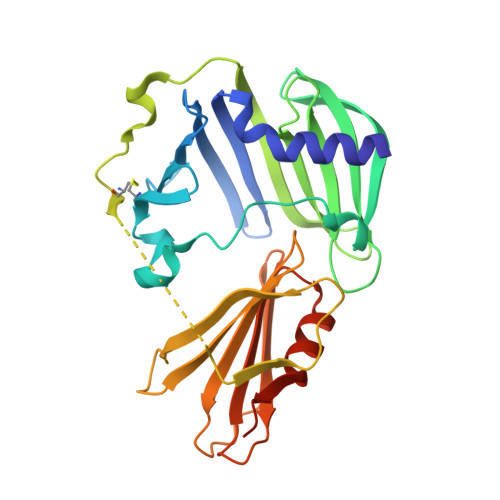
Molecular basis of the lipid-induced MucA-MucB dissociation in Pseudomonas aeruginosa.
Li, T., He, L., Li, C., Kang, M., Song, Y., Zhu, Y., Shen, Y., Zhao, N., Zhao, C., Yang, J., Huang, Q., Mou, X., Tong, A., Yang, J., Wang, Z., Ji, C., Li, H., Tang, H., Bao, R.(2020) Commun Biol 3: 418-418
- PubMed: 32747658
- DOI: https://doi.org/10.1038/s42003-020-01147-1
- Primary Citation of Related Structures:
6JAU - PubMed Abstract:
MucA and MucB are critical negative modulators of sigma factor AlgU and regulate the mucoid conversion of Pseudomonas aeruginosa. Previous studies have revealed that lipid signals antagonize MucA-MucB binding. Here we report the crystal structure of MucB in complex with the periplasmic domain of MucA and polyethylene glycol (PEG), which unveiled an intermediate state preceding the MucA-MucB dissociation. Based on the biochemical experiments, the aliphatic side chain with a polar group was found to be of primary importance for inducing MucA cleavage. These results provide evidence that the hydrophobic cavity of MucB is a primary site for sensing lipid molecules and illustrates the detailed control of conformational switching within MucA-MucB in response to lipophilic effectors.
Organizational Affiliation:
Center of Infectious Diseases, State Key Laboratory of Biotherapy and Cancer Center, West China Hospital, Sichuan University and Collaborative Innovation Center of Biotherapy, Chengdu, China.