Molecular basis for pore blockade of human Na+channel Nav1.2 by the mu-conotoxin KIIIA.
Pan, X., Li, Z., Huang, X., Huang, G., Gao, S., Shen, H., Liu, L., Lei, J., Yan, N.(2019) Science 363: 1309-1313
- PubMed: 30765605
- DOI: https://doi.org/10.1126/science.aaw2999
- Primary Citation of Related Structures:
6J8E - PubMed Abstract:
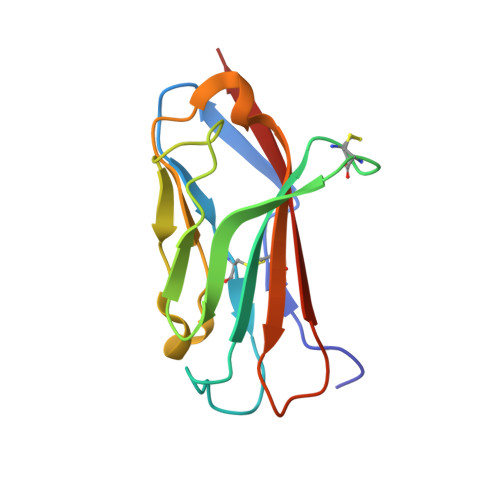
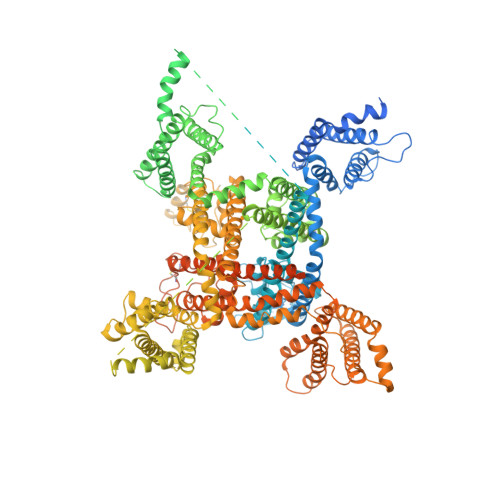
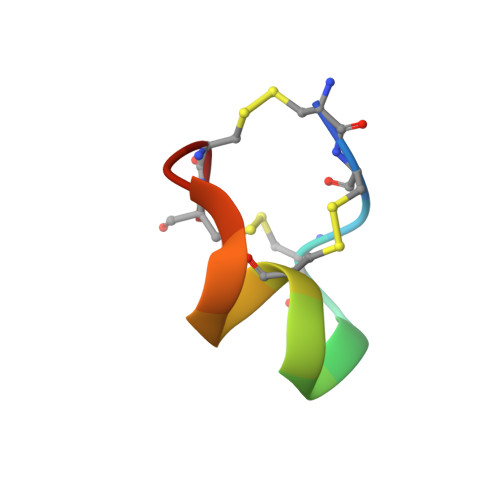
The voltage-gated sodium channel Na v 1.2 is responsible for the initiation and propagation of action potentials in the central nervous system. We report the cryo-electron microscopy structure of human Na v 1.2 bound to a peptidic pore blocker, the μ-conotoxin KIIIA, in the presence of an auxiliary subunit, β2, to an overall resolution of 3.0 angstroms. The immunoglobulin domain of β2 interacts with the shoulder of the pore domain through a disulfide bond. The 16-residue KIIIA interacts with the extracellular segments in repeats I to III, placing Lys 7 at the entrance to the selectivity filter. Many interacting residues are specific to Na v 1.2, revealing a molecular basis for KIIIA specificity. The structure establishes a framework for the rational design of subtype-specific blockers for Na v channels.
Organizational Affiliation:
State Key Laboratory of Membrane Biology, School of Life Sciences and School of Medicine, Tsinghua University, Beijing 100084, China.