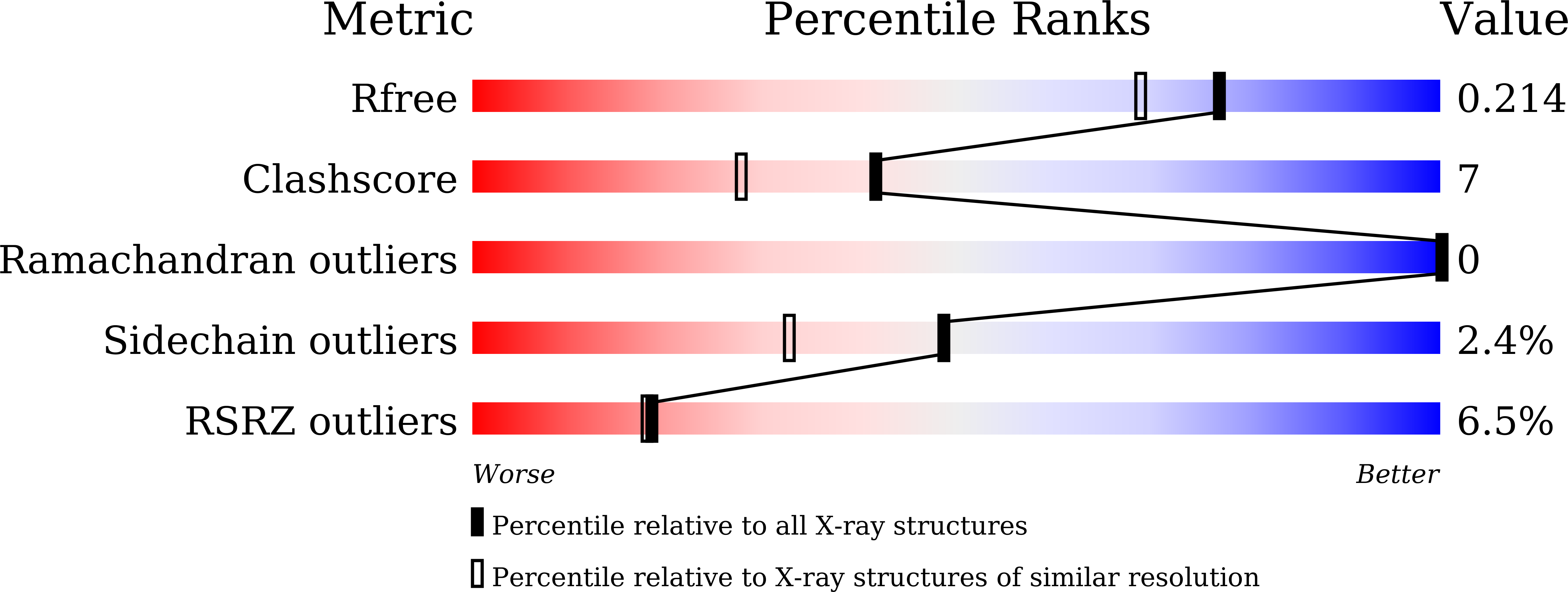
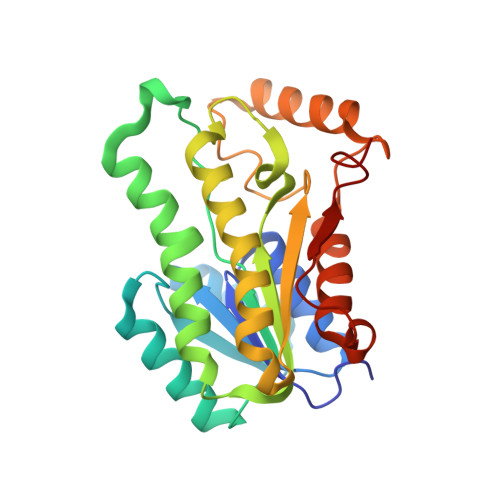
Efficient reductive desymmetrization of bulky 1,3-cyclodiketones enabled by structure-guided directed evolution of a carbonyl reductase.
Chen, X., Zhang, H.L., Ma, M.S., Liu, W., Li, J., Feng, J., Liu, X., Osuna, S., Guo, R.T., Wu, Q., Zhu, D., Ma, Y.(2019) Nat Catal 2: 931-941