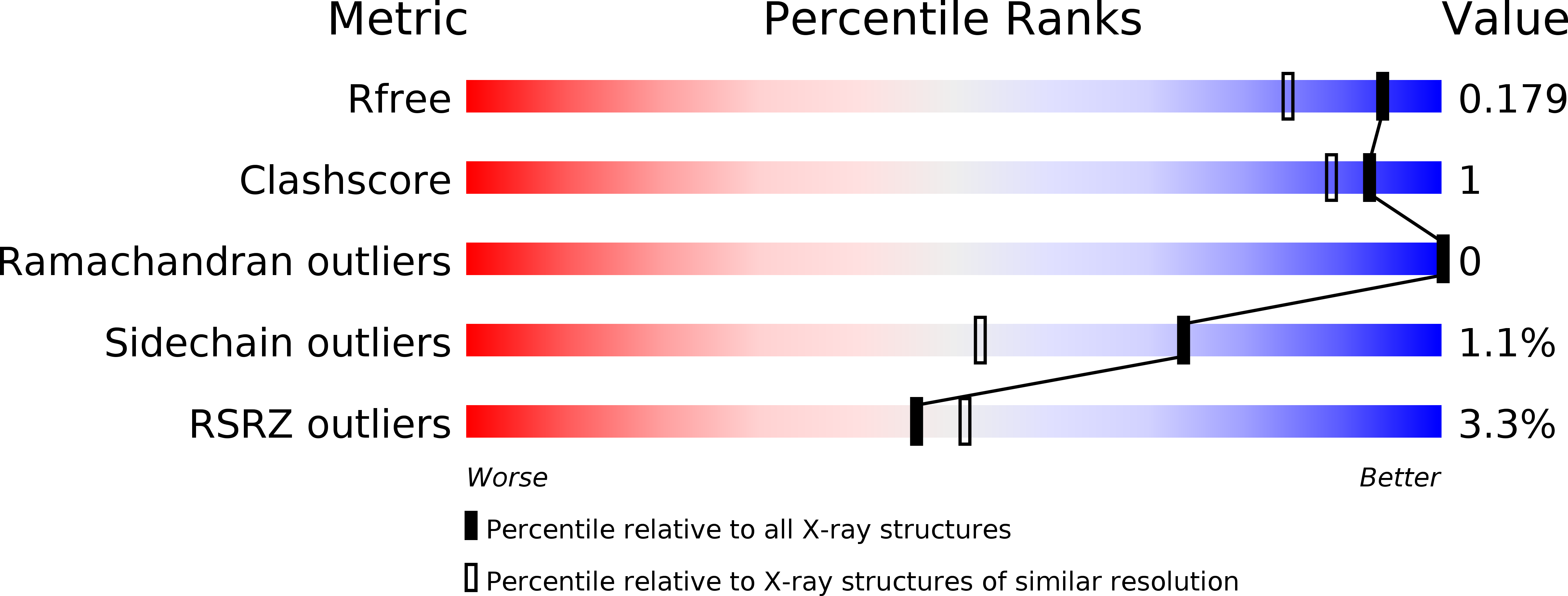
Structural basis for activation of a diguanylate cyclase required for bacterial predation in Bdellovibrio.
Meek, R.W., Cadby, I.T., Moynihan, P.J., Lovering, A.L.(2019) Nat Commun 10: 4086-4086
- PubMed: 31501441
- DOI: https://doi.org/10.1038/s41467-019-12051-6
- Primary Citation of Related Structures:
6HBZ, 6HC0, 6HC1 - PubMed Abstract:
The bacterial second messenger cyclic-di-GMP is a widespread, prominent effector of lifestyle change. An example of this occurs in the predatory bacterium Bdellovibrio bacteriovorus, which cycles between free-living and intraperiplasmic phases after entering (and killing) another bacterium. The initiation of prey invasion is governed by DgcB (GGDEF enzyme) that produces cyclic-di-GMP in response to an unknown stimulus. Here, we report the structure of DgcB, and demonstrate that the GGDEF and sensory forkhead-associated (FHA) domains form an asymmetric dimer. Our structures indicate that the FHA domain is a consensus phosphopeptide sensor, and that the ligand for activation is surprisingly derived from the N-terminal region of DgcB itself. We confirm this hypothesis by determining the structure of a FHA:phosphopeptide complex, from which we design a constitutively-active mutant (confirmed via enzyme assays). Our results provide an understanding of the stimulus driving DgcB-mediated prey invasion and detail a unique mechanism of GGDEF enzyme regulation.
Organizational Affiliation:
Institute for Microbiology and Infection, School of Biosciences, University of Birmingham, Birmingham, B15 2TT, UK.