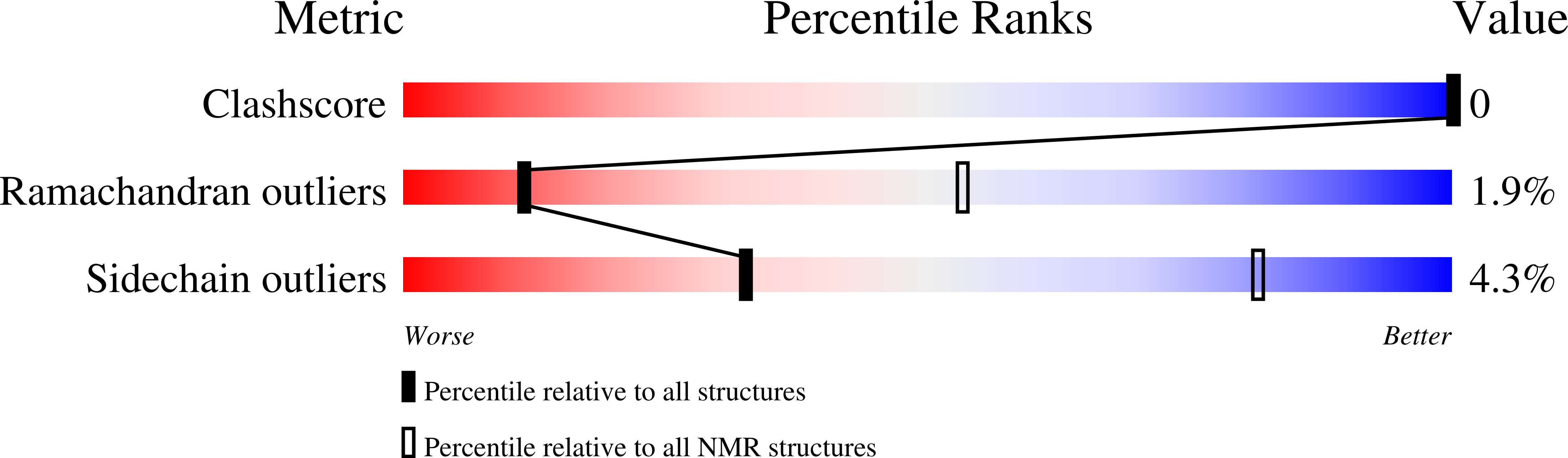From identification to functional characterization of cyriotoxin-1a, an antinociceptive toxin from the spider Cyriopagopus schioedtei.
Goncalves, T.C., Benoit, E., Kurz, M., Lucarain, L., Fouconnier, S., Combemale, S., Jaquillard, L., Schombert, B., Chambard, J.M., Boukaiba, R., Hessler, G., Bohme, A., Bialy, L., Hourcade, S., Beroud, R., De Waard, M., Servent, D., Partiseti, M.(2019) Br J Pharmacol 176: 1298-1314
- PubMed: 30784059
- DOI: https://doi.org/10.1111/bph.14628
- Primary Citation of Related Structures:
6GFT - PubMed Abstract:
The Na V 1.7 channel is highly expressed in dorsal root ganglia of the sensory nervous system and plays a central role in the pain signalling process. We investigated a library prepared from original venoms of 117 different animals to identify new selective inhibitors of this target.
Organizational Affiliation:
Integrated Drug Discovery-High Content Biology, Sanofi R&D, Vitry-sur-Seine, France.