Amelioration of PXR-mediated CYP3A4 induction by mGluR2 modulators.
Vaz, R.J., Li, Y., Chellaraj, V., Reiling, S., Kuntzweiler, T., Yang, D., Shen, H., Batchelor, J.D., Zhang, Y., Chen, X., McLean, L.R., Kosley Jr., R.(2018) Bioorg Med Chem Lett 28: 3194-3196
- PubMed: 30146095
- DOI: https://doi.org/10.1016/j.bmcl.2018.08.022
- Primary Citation of Related Structures:
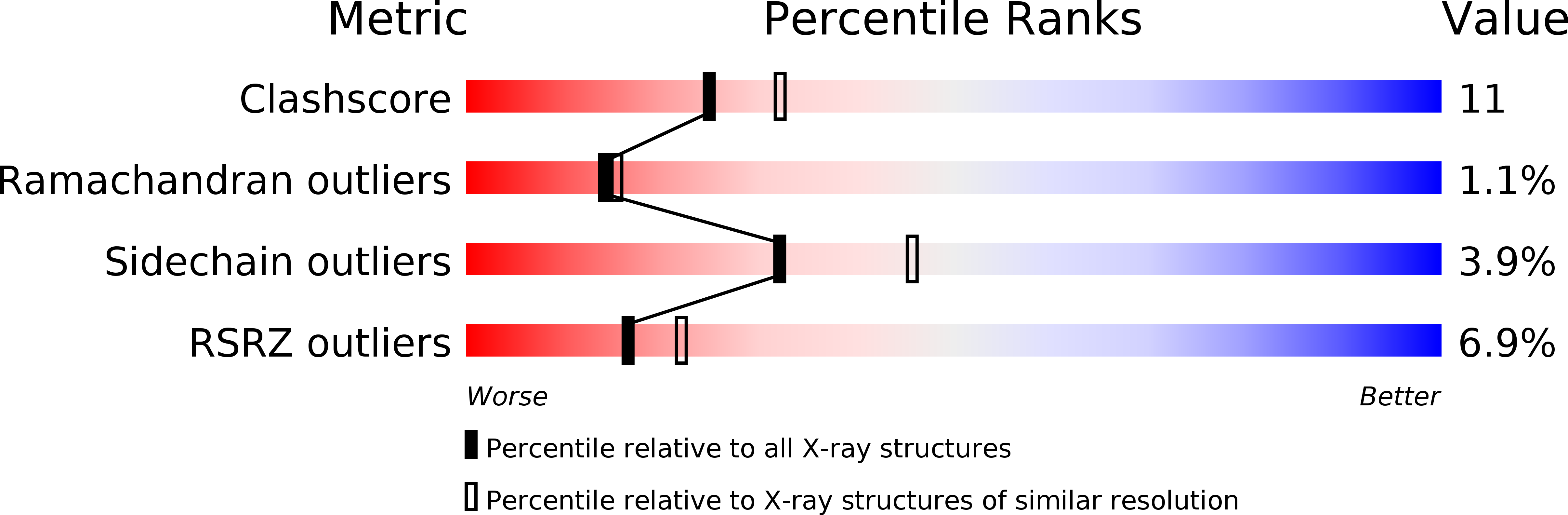
6DUP - PubMed Abstract:
This work describes the rational amelioration of Cytochrome P450 4/5 (CYP3A4/5) induction through the Pregnane-X Receptor (PXR) pathway in a series of compounds that modulate the metabotropic glutamate Receptor 2 (mGluR2) via an allosteric mechanism. The compounds were initially shown to induce CYP3A4/5 via the gold-standard induction assay measured in primary human hepatocytes. This was followed up by testing the compounds in a PXR assay which correlated well with the assay in primary cells. Further, one of the compounds was crystallized with PXR (pdb code 6DUP). Analysis of this co-crystal structure, together with previously published PXR co-crystal structures, lead to modification ideas. The compounds synthesized based on these ideas were shown not to be CYP3A4/5 inducers. The mGluR2 activity of the resulting compounds was maintained.
Organizational Affiliation:
Integrated Drug Discovery, Sanofi US, 153-2nd Ave., Waltham, MA 02451, USA. Electronic address: roy.vaz@sanofi.com.