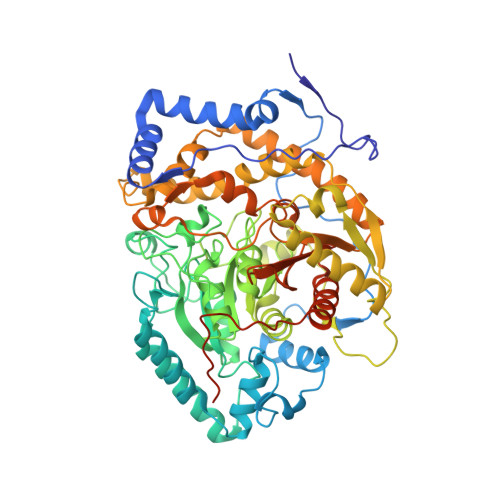
Structural analysis of a plant fatty acid amide hydrolase provides insights into the evolutionary diversity of bioactive acylethanolamides.
Aziz, M., Wang, X., Tripathi, A., Bankaitis, V.A., Chapman, K.D.(2019) J Biol Chem 294: 7419-7432
- PubMed: 30894416
- DOI: https://doi.org/10.1074/jbc.RA118.006672
- Primary Citation of Related Structures:
6DHV, 6DII - PubMed Abstract:
N -Acylethanolamines (NAEs) are fatty acid derivatives that in animal systems include the well-known bioactive metabolites of the endocannabinoid signaling pathway. Plants use NAE signaling as well, and these bioactive molecules often have oxygenated acyl moieties. Here, we report the three-dimensional crystal structures of the signal-terminating enzyme fatty acid amide hydrolase (FAAH) from Arabidopsis in its apo and ligand-bound forms at 2.1- and 3.2-Å resolutions, respectively. This plant FAAH structure revealed features distinct from those of the only other available FAAH structure (rat). The structures disclosed that although catalytic residues are conserved with the mammalian enzyme, AtFAAH has a more open substrate-binding pocket that is partially lined with polar residues. Fundamental differences in the organization of the membrane-binding "cap" and the membrane access channel also were evident. In accordance with the observed structural features of the substrate-binding pocket, kinetic analysis showed that AtFAAH efficiently uses both unsubstituted and oxygenated acylethanolamides as substrates. Moreover, comparison of the apo and ligand-bound AtFAAH structures identified three discrete sets of conformational changes that accompany ligand binding, suggesting a unique "squeeze and lock" substrate-binding mechanism. Using molecular dynamics simulations, we evaluated these conformational changes further and noted a partial unfolding of a random-coil helix within the region 531-537 in the apo structure but not in the ligand-bound form, indicating that this region likely confers plasticity to the substrate-binding pocket. We conclude that the structural divergence in bioactive acylethanolamides in plants is reflected in part in the structural and functional properties of plant FAAHs.
Organizational Affiliation:
From the BioDiscovery Institute and Department of Biological Sciences, University of North Texas, Denton, Texas 76203 and.