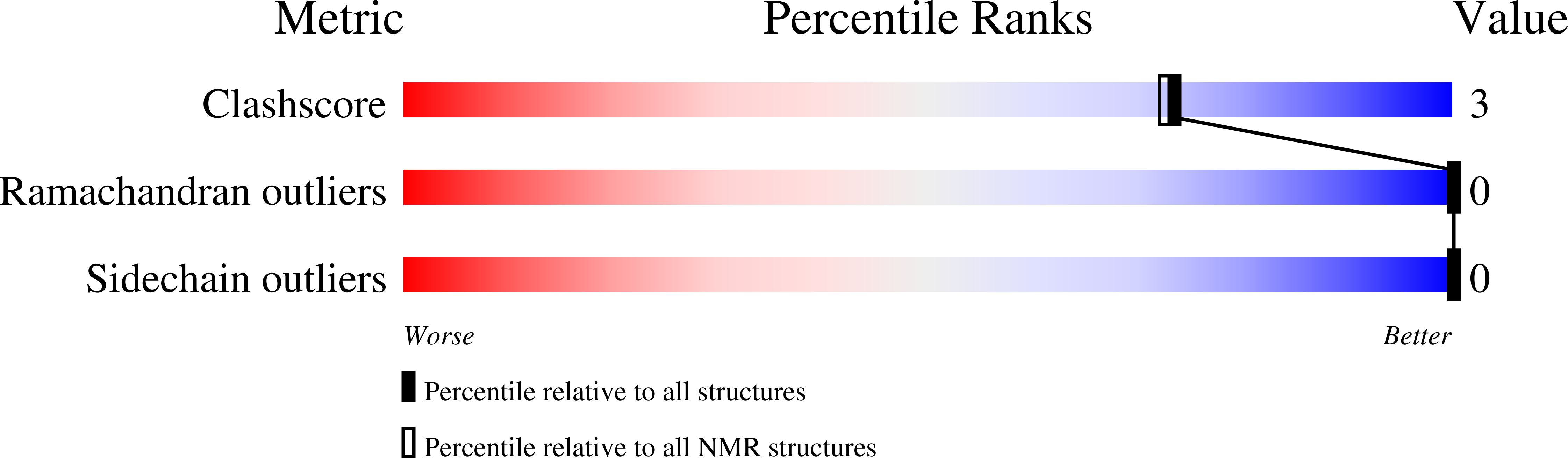Discovery and Characterization of Cyclotides from Rinorea Species.
Niyomploy, P., Chan, L.Y., Harvey, P.J., Poth, A.G., Colgrave, M.L., Craik, D.J.(2018) J Nat Prod 81: 2512-2520
- PubMed: 30387611
- DOI: https://doi.org/10.1021/acs.jnatprod.8b00572
- Primary Citation of Related Structures:
6DHR - PubMed Abstract:
Cyclotides are macrocyclic cystine-knotted peptides most commonly found in the Violaceae plant family. Although Rinorea is the second-largest genera within the Violaceae family, few studies have examined whether or not they contain cyclotides. To further our understanding of cyclotide diversity and evolution, we examined the cyclotide content of two Rinorea species found in Southeast Asia: R. virgata and R. bengalensis. Seven cyclotides were isolated from R. virgata (named Rivi1-7), and a known cyclotide (cT10) was found in R. bengalensis. Loops 2, 5, and 6 of Rivi1-4 contained sequences not previously seen in corresponding loops of known cyclotides, thereby expanding our understanding of the diversity of cyclotides. In addition, the sequence of loop 2 of Rivi3 and Rivi4 were identical to some related noncyclic "acyclotides" from the Poaceae plant family. As only acyclotides, but not cyclotides, have been reported in monocotyledons thus far, our findings support an evolutionary link between monocotyledon-derived ancestral cyclotide precursors and dicotyledon-derived cyclotides. Furthermore, Rivi2 and Rivi3 had comparable cytotoxic activities to the most cytotoxic cyclotide known to date: cycloviolacin O2 from Viola odorata; yet, unlike cycloviolacin O2, they did not show hemolytic activity. Therefore, these cyclotides represent novel scaffolds for use in future anticancer drug design.
Organizational Affiliation:
Institute for Molecular Bioscience , The University of Queensland , Brisbane , Queensland 4072 , Australia.