The N-terminal domains of FLASH and Lsm11 form a 2:1 heterotrimer for histone pre-mRNA 3'-end processing.
Aik, W.S., Lin, M.H., Tan, D., Tripathy, A., Marzluff, W.F., Dominski, Z., Chou, C.Y., Tong, L.(2017) PLoS One 12: e0186034-e0186034
- PubMed: 29020104
- DOI: https://doi.org/10.1371/journal.pone.0186034
- Primary Citation of Related Structures:
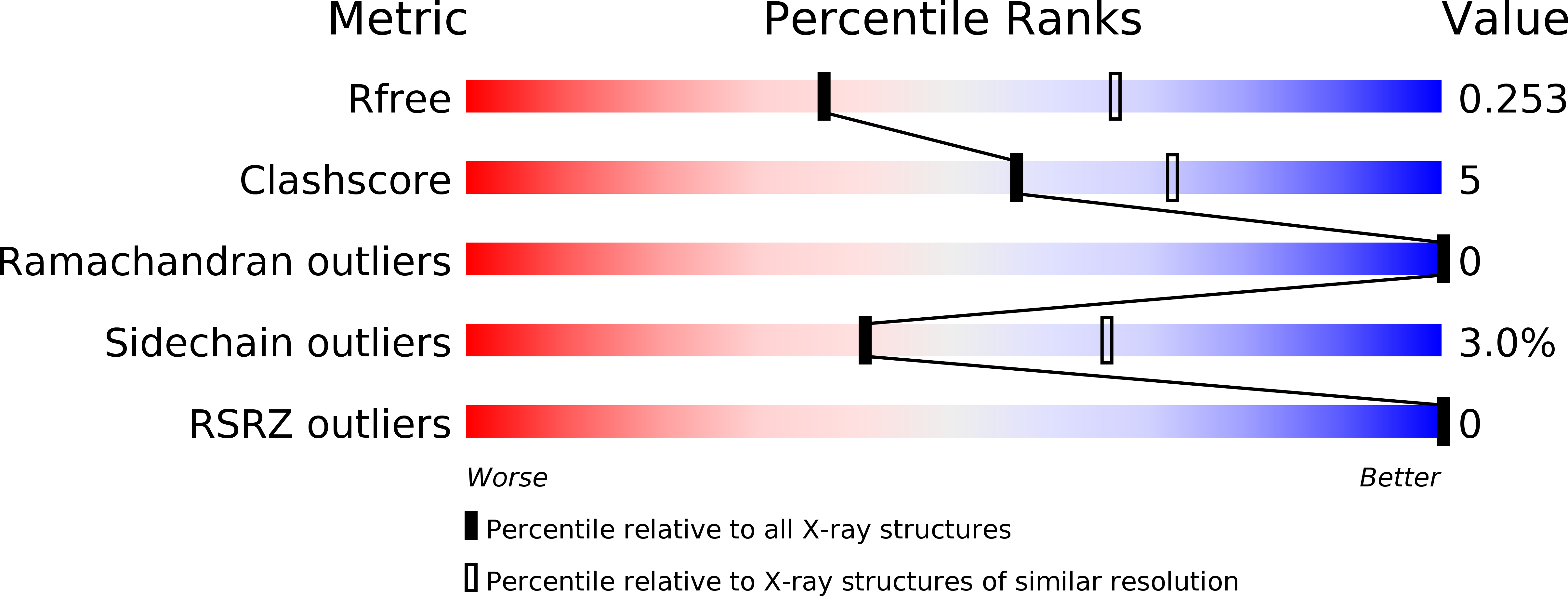
6ANO, 6AOZ, 6AP0 - PubMed Abstract:
Unlike canonical pre-mRNAs, animal replication-dependent histone pre-mRNAs lack introns and are processed at the 3'-end by a mechanism distinct from cleavage and polyadenylation. They have a 3' stem loop and histone downstream element (HDE) that are recognized by stem-loop binding protein (SLBP) and U7 snRNP, respectively. The N-terminal domain (NTD) of Lsm11, a component of U7 snRNP, interacts with FLASH NTD and these two proteins recruit the histone cleavage complex containing the CPSF-73 endonuclease for the cleavage reaction. Here, we determined crystal structures of FLASH NTD and found that it forms a coiled-coil dimer. Using solution light scattering, we characterized the stoichiometry of the FLASH NTD-Lsm11 NTD complex and found that it is a 2:1 heterotrimer, which is supported by observations from analytical ultracentrifugation and crosslinking.
Organizational Affiliation:
Department of Biological Sciences, Columbia University, New York, New York, United States of America.