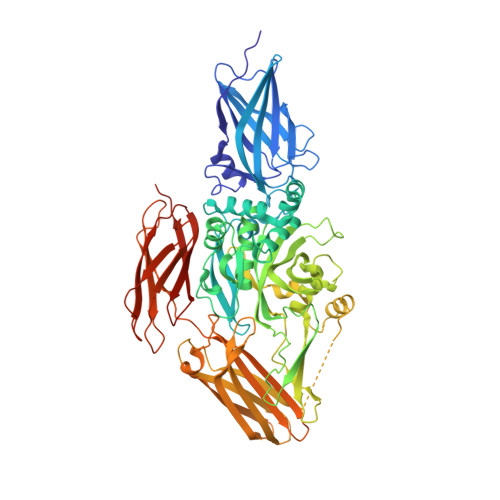
Structure of natural variant transglutaminase 2 reveals molecular basis of gaining stability and higher activity.
Ha, H.J., Kwon, S., Jeong, E.M., Kim, C.M., Lee, K.B., Kim, I.G., Park, H.H.(2018) PLoS One 13: e0204707-e0204707
- PubMed: 30321187
- DOI: https://doi.org/10.1371/journal.pone.0204707
- Primary Citation of Related Structures:
6A8P - PubMed Abstract:
Multi-functional transglutaminase 2 (TG2), which possesses protein cross-linking and GTP hydrolysis activities, is involved in various cellular processes, including apoptosis, angiogenesis, wound healing, and neuronal regeneration, and is associated with many human diseases, including inflammatory disease, celiac disease, neurodegenerative disease, diabetes, tissue fibrosis, and cancers. Although most biochemical and cellular studies have been conducted with the TG2 (G224) form, the TG2 (G224V) form has recently emerged as a putative natural variant of TG2. In this study, we characterized the putative natural form of TG2, TG2 (G224V), and through a new crystal structure of TG2 (G224V), we revealed how TG2 (G224V) gained stability and higher Ca2+-dependent activity than an artificial variant of TG2 (G224).
Organizational Affiliation:
School of Pharmacy, Chung-Ang University, Seoul, South Korea.