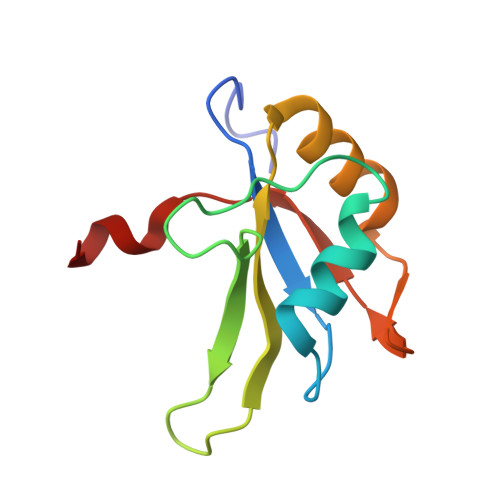
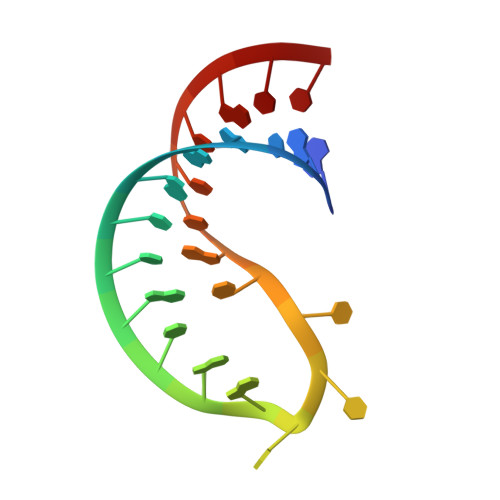
The Solution Structure of FUS Bound to RNA Reveals a Bipartite Mode of RNA Recognition with Both Sequence and Shape Specificity.
Loughlin, F.E., Lukavsky, P.J., Kazeeva, T., Reber, S., Hock, E.M., Colombo, M., Von Schroetter, C., Pauli, P., Clery, A., Muhlemann, O., Polymenidou, M., Ruepp, M.D., Allain, F.H.(2019) Mol Cell 73: 490-504.e6
- PubMed: 30581145
- DOI: https://doi.org/10.1016/j.molcel.2018.11.012
- Primary Citation of Related Structures:
6G99, 6GBM - PubMed Abstract:
Fused in sarcoma (FUS) is an RNA binding protein involved in regulating many aspects of RNA processing and linked to several neurodegenerative diseases. Transcriptomics studies indicate that FUS binds a large variety of RNA motifs, suggesting that FUS RNA binding might be quite complex. Here, we present solution structures of FUS zinc finger (ZnF) and RNA recognition motif (RRM) domains bound to RNA. These structures show a bipartite binding mode of FUS comprising of sequence-specific recognition of a NGGU motif via the ZnF and an unusual shape recognition of a stem-loop RNA via the RRM. In addition, sequence-independent interactions via the RGG repeats significantly increase binding affinity and promote destabilization of structured RNA conformation, enabling additional binding. We further show that disruption of the RRM and ZnF domains abolishes FUS function in splicing. Altogether, our results rationalize why deciphering the RNA binding mode of FUS has been so challenging.
Organizational Affiliation:
Department of Biology, Institute of Molecular Biology and Biophysics, ETH Zürich, 8093 Zürich, Switzerland; Monash Biomedicine Discovery Institute, Department of Biochemistry and Molecular Biology, Monash University, Clayton, 3800 VIC, Australia. Electronic address: fionna.loughlin@monash.edu.