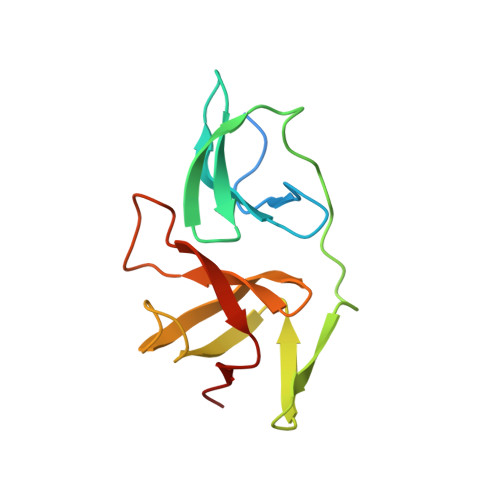
Structure and nucleic acid binding properties of KOW domains 4 and 6-7 of human transcription elongation factor DSIF.
Zuber, P.K., Hahn, L., Reinl, A., Schweimer, K., Knauer, S.H., Gottesman, M.E., Rosch, P., Wohrl, B.M.(2018) Sci Rep 8: 11660-11660
- PubMed: 30076330
- DOI: https://doi.org/10.1038/s41598-018-30042-3
- Primary Citation of Related Structures:
6EQY, 6ER0 - PubMed Abstract:
The human transcription elongation factor DSIF is highly conserved throughout all kingdoms of life and plays multiple roles during transcription. DSIF is a heterodimer, consisting of Spt4 and Spt5 that interacts with RNA polymerase II (RNAP II). DSIF binds to the elongation complex and induces promoter-proximal pausing of RNAP II. Human Spt5 consists of a NusG N-terminal (NGN) domain motif, which is followed by several KOW domains. We determined the solution structures of the human Spt5 KOW4 and the C-terminal domain by nuclear magnetic resonance spectroscopy. In addition to the typical KOW fold, the solution structure of KOW4 revealed an N-terminal four-stranded β-sheet, previously designated as the KOW3-KOW4 linker. In solution, the C-terminus of Spt5 consists of two β-barrel folds typical for KOW domains, designated KOW6 and KOW7. We also analysed the nucleic acid and RNAP II binding properties of the KOW domains. KOW4 variants interacted with nucleic acids, preferentially single stranded RNA, whereas no nucleic acid binding could be detected for KOW6-7. Weak binding of KOW4 to the RNAP II stalk, which is comprised of Rpb4/7, was also detected, consistent with transient interactions between Spt5 and these RNAP II subunits.
Organizational Affiliation:
Universität Bayreuth, Lehrstuhl Biopolymere, Universitätsstr. 30, D-95447, Bayreuth, Germany.