Crystal Structure of the Marburg Virus Nucleoprotein Core Domain Chaperoned by a VP35 Peptide Reveals a Conserved Drug Target for Filovirus
Zhu, T., Song, H., Peng, R., Shi, Y., Qi, J., Gao, G.F.(2017) J Virol 91
- PubMed: 28659479
- DOI: https://doi.org/10.1128/JVI.00996-17
- Primary Citation of Related Structures:
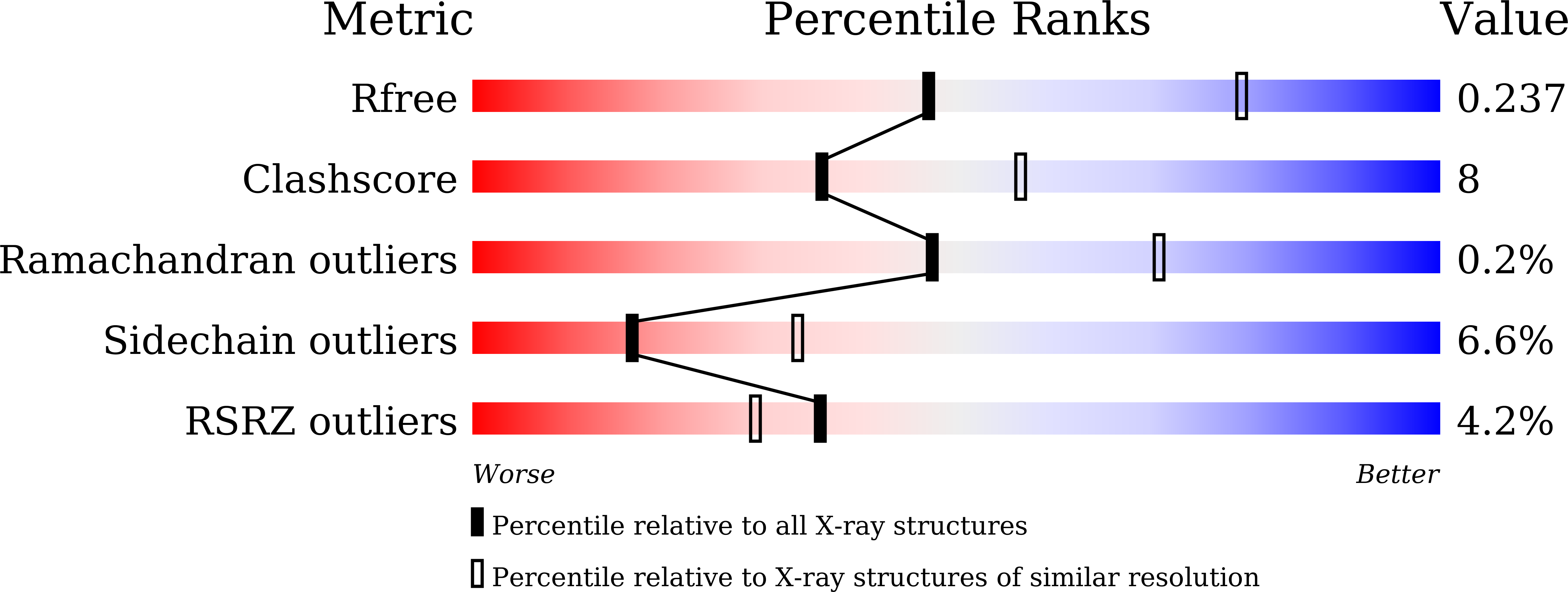
5XSQ - PubMed Abstract:
Filovirus nucleoprotein (NP), viral protein 35 (VP35), and polymerase L are essential for viral replication and nucleocapsid formation. Here, we identify a 28-residue peptide (NP binding peptide [NPBP]) from Marburg virus (MARV) VP35 through sequence alignment with previously identified Ebola virus (EBOV) NPBP, which bound to the core region (residues 18 to 344) of the N-terminal portion of MARV NP with high affinity. The crystal structure of the MARV NP core/NPBP complex at a resolution of 2.6 Å revealed that NPBP binds to the C-terminal region of the NP core via electrostatic and nonpolar interactions. Further structural analysis revealed that the MARV and EBOV NP cores hold a conserved binding pocket for NPBP, and this pocket could serve as a promising target for the design of universal drugs against filovirus infection. In addition, cross-binding assays confirmed that the NP core of MARV or EBOV can bind the NPBP from the other virus, although with moderately reduced binding affinities that result from termini that are distinct between the MARV and EBOV NPBPs. IMPORTANCE Historically, Marburg virus (MARV) has caused severe disease with up to 90% lethality. Among the viral proteins produced by MARV, NP and VP35 are both multifunctional proteins that are essential for viral replication. In its relative, Ebola virus (EBOV), an N-terminal peptide from VP35 binds to the NP N-terminal region with high affinity. Whether this is a common mechanism among filoviruses is an unsolved question. Here, we present the crystal structure of a complex that consists of the core domain of MARV NP and the NPBP peptide from VP35. As we compared MARV NPBP with EBOV NPBP, several different features at the termini were identified. Although these differences reduce the affinity of the NP core for NPBPs across genera, a conserved pocket in the C-terminal region of the NP core makes cross-species binding possible. Our results expand our knowledge of filovirus NP-VP35 interactions and provide more details for therapeutic intervention.
Organizational Affiliation:
Research Network of Immunity and Health (RNIH), Beijing Institutes of Life Science, Chinese Academy of Sciences, Beijing, China.