New Insights into PI3K Inhibitor Design using X-ray Structures of PI3K alpha Complexed with a Potent Lead Compound.
Yang, X., Zhang, X., Huang, M., Song, K., Li, X., Huang, M., Meng, L., Zhang, J.(2017) Sci Rep 7: 14572-14572
- PubMed: 29109464
- DOI: https://doi.org/10.1038/s41598-017-15260-5
- Primary Citation of Related Structures:
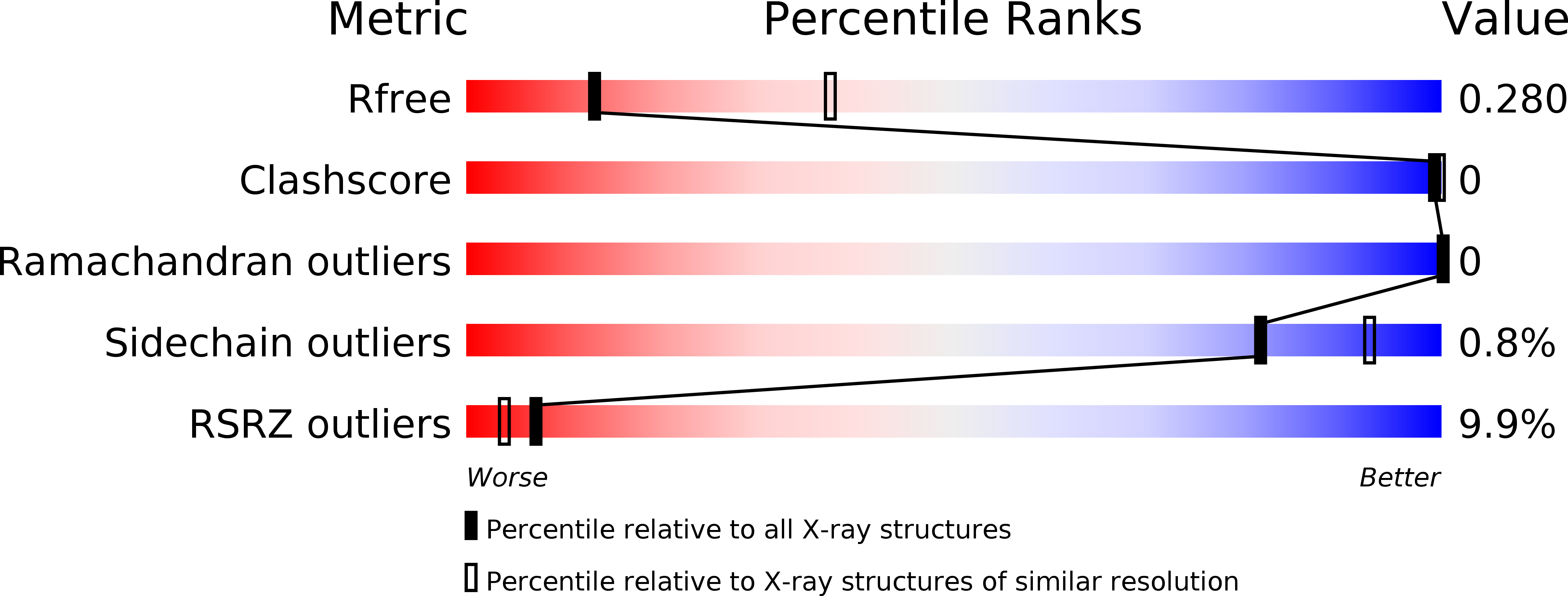
5XGH - PubMed Abstract:
Phosphatidylinositol 3-kinase α is an attractive target to potentially treat a range of cancers. Herein, we described the evolution of a reported PI3K inhibitor into a moderate PI3Kα inhibitor with a low molecular weight. We used X-ray crystallography to describe the accurate binding mode of the compound YXY-4F. A comparison of the p110α-YXY-4F and apo p110α complexes showed that YXY-4F induced additional space by promoting a flexible conformational change in residues Ser773 and Ser774 in the PI3Kα ATP catalytic site. Specifically, residue 773(S) in PI3Kα is quite different from that of PI3Kβ (D), γ (A), and δ (D), which might guide further optimization of substituents around the NH group and phenyl group to improve the selectivity and potency of PI3Kα.
Organizational Affiliation:
Institute of Bioinformatics and Medical Engineering, School of Electrical and Information Engineering, Jiangsu University of Technology, Changzhou, 213001, China.