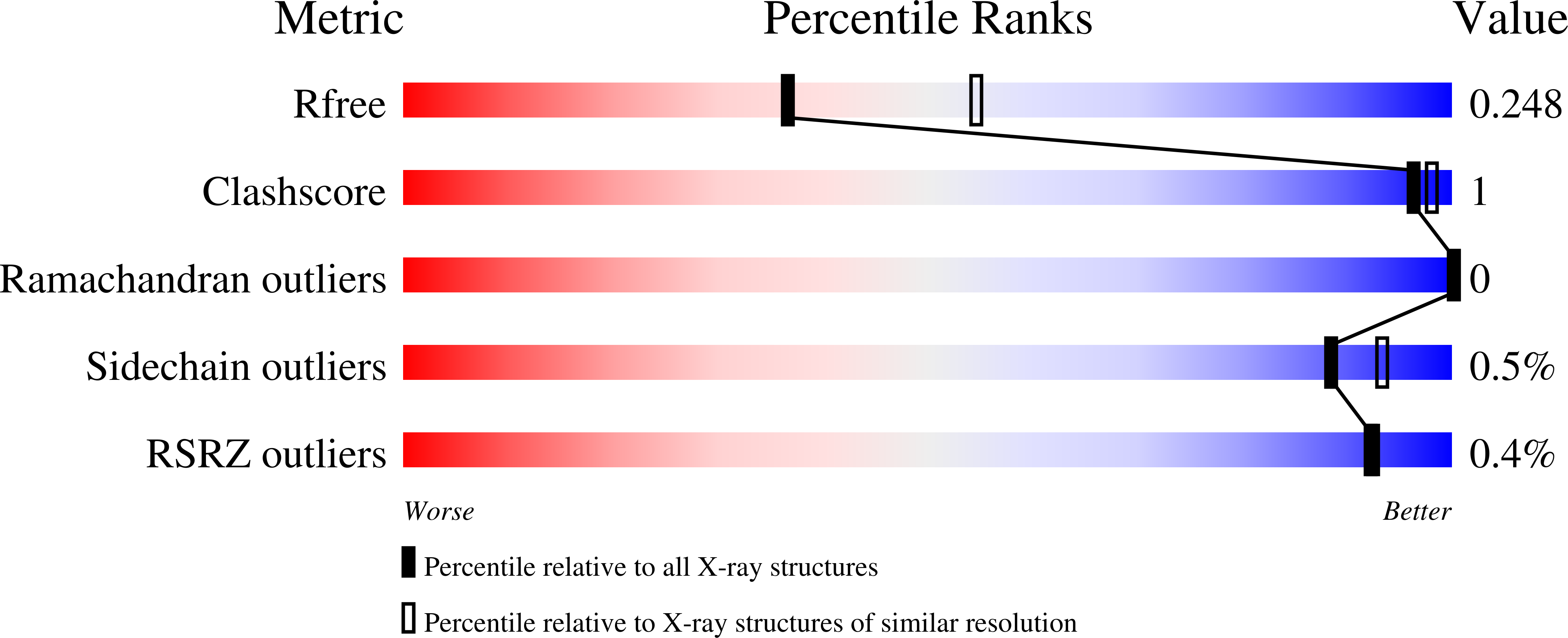
Structural and Functional Characterization of the Histidine Phosphatase Domains of Human Sts-1 and Sts-2.
Zhou, W., Yin, Y., Weinheimer, A.S., Kaur, N., Carpino, N., French, J.B.(2017) Biochemistry 56: 4637-4645
- PubMed: 28759203
- DOI: https://doi.org/10.1021/acs.biochem.7b00638
- Primary Citation of Related Structures:
5VR6, 5W5G, 5WDI - PubMed Abstract:
The suppressor of T cell signaling (Sts) proteins, Sts-1 and Sts-2, are homologous phosphatases that negatively regulate signaling pathways downstream of the T cell receptor. Functional inactivation of Sts-1 and Sts-2 in a murine model leads to resistance to systemic infection by the opportunistic pathogen, Candida albicans. This suggests that modulation of the host immune response by inhibiting Sts function may be a viable strategy for treating these deadly fungal pathogen infections. To better understand the molecular determinants of function and structure, we characterized the structure and steady-state kinetics of the histidine phosphatase domains of human Sts-1 (Sts-1 HP ) and Sts-2 (Sts-2 HP ). We determined the X-ray crystal structures of unliganded Sts-1 HP and Sts-1 HP in complex with sulfate to 2.5 and 1.9 Å, respectively, and the structure of Sts-2 HP with sulfate to 2.4 Å. The steady-state kinetic analysis shows, as expected, that Sts-1 HP has a phosphatase activity significantly higher than that of Sts-2 HP and that the human and mouse proteins behave similarly. In addition, comparison of the phosphatase activity of full-length Sts-1 protein to Sts-1 HP reveals similar kinetics, indicating that Sts-1 HP is a functional surrogate for the native protein. We also tested known phosphatase inhibitors and determined that the SHP-1 inhibitor, PHPS1, is a potent inhibitor of Sts-1 (K i = 1.05 ± 0.15 μM). Finally, we demonstrated that human Sts-1 has robust phosphatase activity against the substrate, Zap-70, in a cell-based assay. Collectively, these data suggest that the human Sts proteins are druggable targets and provide a structural basis for future drug development efforts.
Organizational Affiliation:
Department of Chemistry, Stony Brook University , Stony Brook, New York 11794, United States.