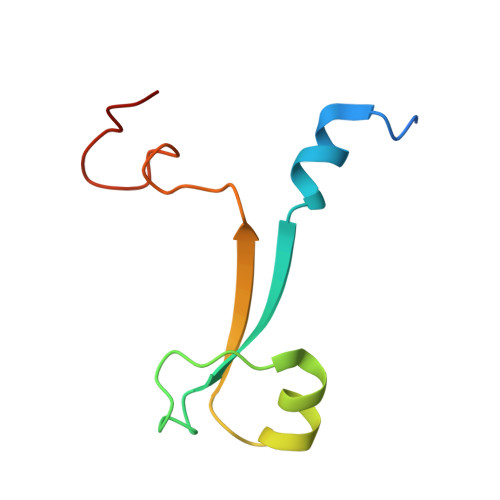
Crystal structure of the UBR-box from UBR6/FBXO11 reveals domain swapping mediated by zinc binding.
Munoz-Escobar, J., Kozlov, G., Gehring, K.(2017) Protein Sci 26: 2092-2097
- PubMed: 28691247
- DOI: https://doi.org/10.1002/pro.3227
- Primary Citation of Related Structures:
5VMD - PubMed Abstract:
The UBR-box is a 70-residue zinc finger domain present in the UBR family of E3 ubiquitin ligases that directly binds N-terminal degradation signals in substrate proteins. UBR6, also called FBXO11, is an UBR-box containing E3 ubiquitin ligase that does not bind N-terminal signals. Here, we present the crystal structure of the UBR-box domain from human UBR6. The dimeric crystal structure reveals a unique form of domain swapping mediated by zinc coordination, where three independent protein chains come together to regenerate the topology of the monomeric UBR-box fold. Analysis of the structure suggests that the absence of N-terminal residue binding arises from the lack of an amino acid binding pocket.
Organizational Affiliation:
Department of Biochemistry, Groupe de Recherche Axé sur la Structure des Protéines, McGill University, Montreal, Quebec, H3G0B1, Canada.