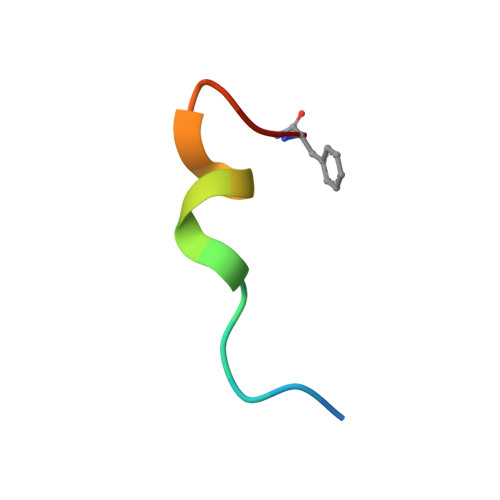
Probing the Mechanism of Receptor Activity-Modifying Protein Modulation of GPCR Ligand Selectivity through Rational Design of Potent Adrenomedullin and Calcitonin Gene-Related Peptide Antagonists.
Booe, J.M., Warner, M.L., Roehrkasse, A.M., Hay, D.L., Pioszak, A.A.(2018) Mol Pharmacol 93: 355-367
- PubMed: 29363552
- DOI: https://doi.org/10.1124/mol.117.110916
- Primary Citation of Related Structures:
5V6Y - PubMed Abstract:
Binding of the vasodilator peptides adrenomedullin (AM) and calcitonin gene-related peptide (CGRP) to the class B G protein-coupled receptor calcitonin receptor-like receptor (CLR) is modulated by receptor activity-modifying proteins (RAMPs). RAMP1 favors CGRP, whereas RAMP2 and RAMP3 favor AM. Crystal structures of peptide-bound RAMP1/2-CLR extracellular domain (ECD) heterodimers suggested RAMPs alter ligand preference through direct peptide contacts and allosteric modulation of CLR. Here, we probed this dual mechanism through rational structure-guided design of AM and CGRP antagonist variants. Variants were characterized for binding to purified RAMP1/2-CLR ECD and for antagonism of the full-length CGRP (RAMP1:CLR), AM 1 (RAMP2:CLR), and AM 2 (RAMP3:CLR) receptors. Short nanomolar affinity AM(37-52) and CGRP(27-37) variants were obtained through substitutions including AM S45W/Q50W and CGRP K35W/A36S designed to stabilize their β -turn. K46L and Y52F substitutions designed to exploit RAMP allosteric effects and direct peptide contacts, respectively, yielded AM variants with selectivity for the CGRP receptor over the AM 1 receptor. AM(37-52) S45W/K46L/Q50W/Y52F exhibited nanomolar potency at the CGRP receptor and micromolar potency at AM 1 A 2.8-Å resolution crystal structure of this variant bound to the RAMP1-CLR ECD confirmed that it bound as designed. CGRP(27-37) N31D/S34P/K35W/A36S exhibited potency and selectivity comparable to the traditional antagonist CGRP(8-37). Giving this variant the ability to contact RAMP2 through the F37Y substitution increased affinity for AM 1 , but it still preferred the CGRP receptor. These potent peptide antagonists with altered selectivity inform the development of AM/CGRP-based pharmacological tools and support the hypothesis that RAMPs alter CLR ligand selectivity through allosteric effects and direct peptide contacts.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, University of Oklahoma Health Sciences Center, Oklahoma City, Oklahoma (J.M.B., M.L.W., A.M.R., A.A.P.) and School of Biological Sciences, University of Auckland, Auckland, New Zealand (D.L.H.).