Small molecule inhibitors and CRISPR/Cas9 mutagenesis demonstrate that SMYD2 and SMYD3 activity are dispensable for autonomous cancer cell proliferation.
Thomenius, M.J., Totman, J., Harvey, D., Mitchell, L.H., Riera, T.V., Cosmopoulos, K., Grassian, A.R., Klaus, C., Foley, M., Admirand, E.A., Jahic, H., Majer, C., Wigle, T., Jacques, S.L., Gureasko, J., Brach, D., Lingaraj, T., West, K., Smith, S., Rioux, N., Waters, N.J., Tang, C., Raimondi, A., Munchhof, M., Mills, J.E., Ribich, S., Porter Scott, M., Kuntz, K.W., Janzen, W.P., Moyer, M., Smith, J.J., Chesworth, R., Copeland, R.A., Boriack-Sjodin, P.A.(2018) PLoS One 13: e0197372-e0197372
- PubMed: 29856759
- DOI: https://doi.org/10.1371/journal.pone.0197372
- Primary Citation of Related Structures:
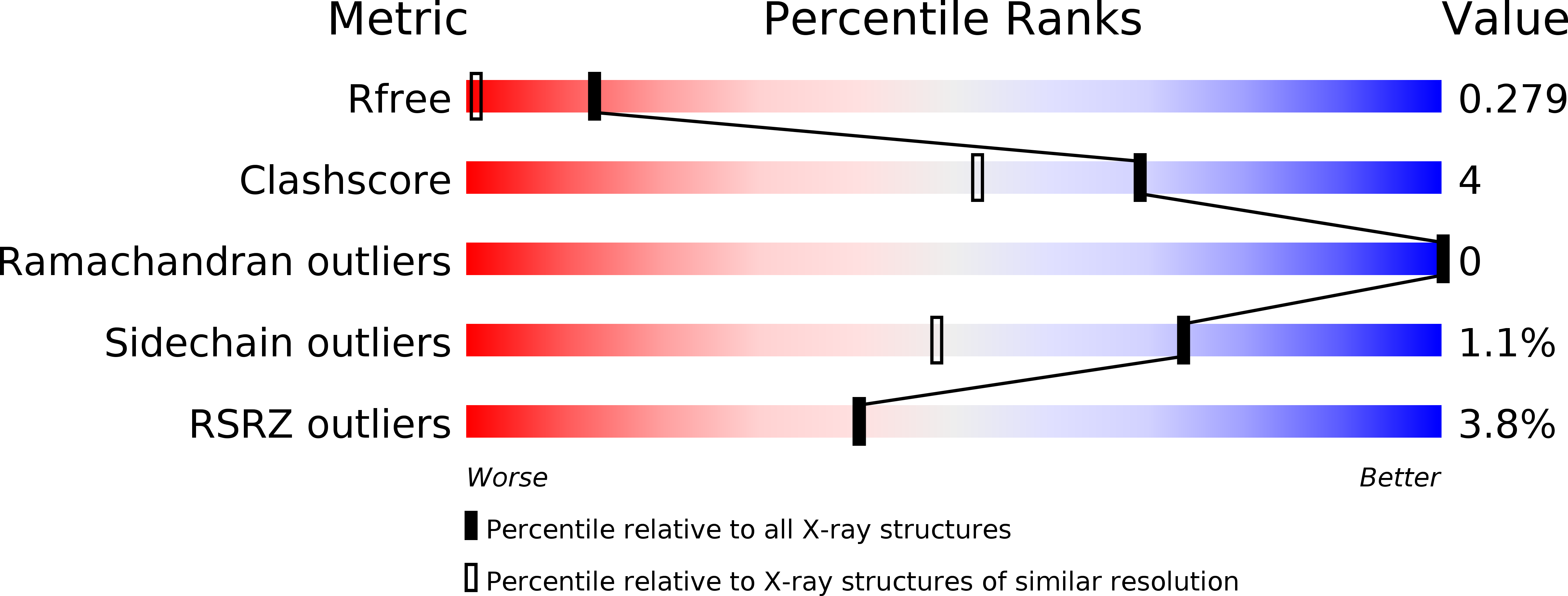
5V37, 5V3H - PubMed Abstract:
A key challenge in the development of precision medicine is defining the phenotypic consequences of pharmacological modulation of specific target macromolecules. To address this issue, a variety of genetic, molecular and chemical tools can be used. All of these approaches can produce misleading results if the specificity of the tools is not well understood and the proper controls are not performed. In this paper we illustrate these general themes by providing detailed studies of small molecule inhibitors of the enzymatic activity of two members of the SMYD branch of the protein lysine methyltransferases, SMYD2 and SMYD3. We show that tool compounds as well as CRISPR/Cas9 fail to reproduce many of the cell proliferation findings associated with SMYD2 and SMYD3 inhibition previously obtained with RNAi based approaches and with early stage chemical probes.
Organizational Affiliation:
Epizyme, Inc., Cambridge, Massachusetts, United States of America.