Structural, biochemical and biophysical characterization of recombinant human fumarate hydratase.
Ajalla Aleixo, M.A., Rangel, V.L., Rustiguel, J.K., de Padua, R.A.P., Nonato, M.C.(2019) FEBS J
- PubMed: 30761759
- DOI: https://doi.org/10.1111/febs.14782
- Primary Citation of Related Structures:
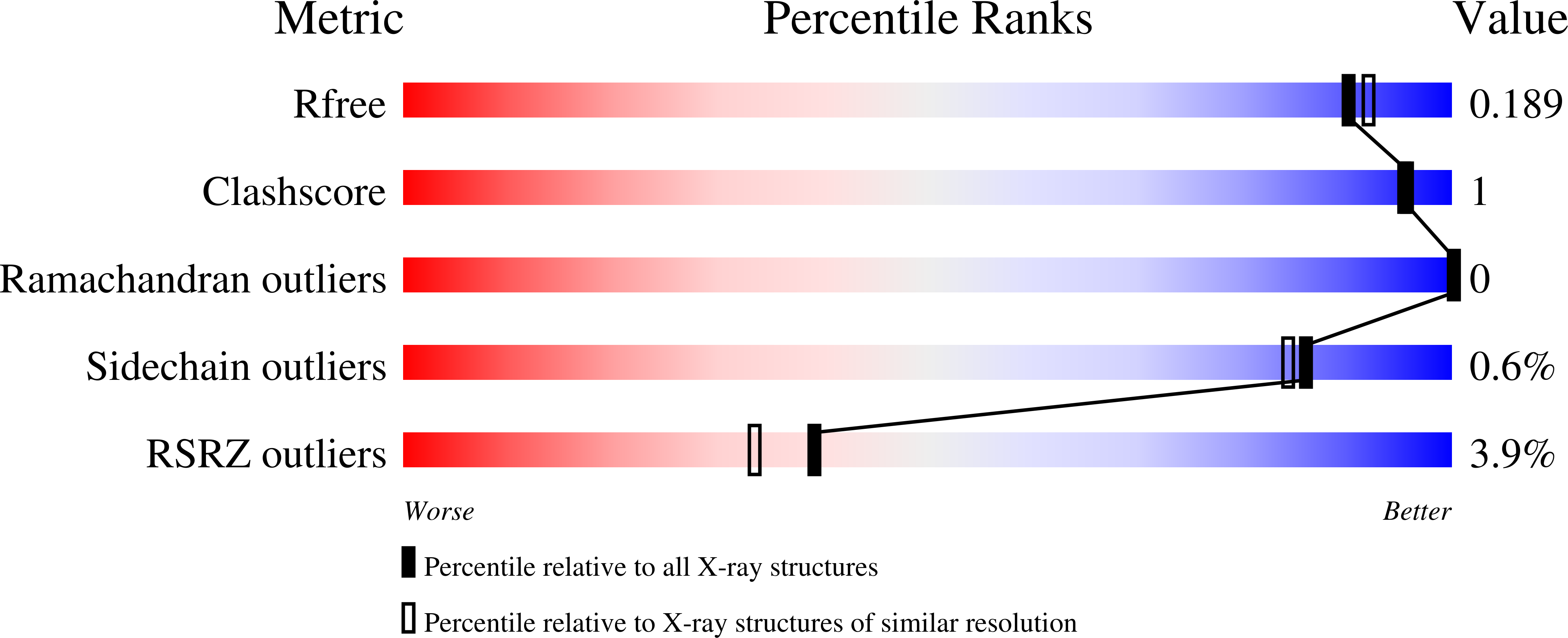
5UPP - PubMed Abstract:
Fumarate hydratases (FHs, fumarases) catalyze the reversible conversion of fumarate into l-malate. FHs are distributed over all organisms and play important roles in energy production, DNA repair and as tumor suppressors. They are very important targets both in the study of human metabolic disorders and as potential therapeutic targets in neglected tropical diseases and tuberculosis. In this study, human FH (HsFH) was characterized by using enzyme kinetics, differential scanning fluorimetry and X-ray crystallography. For the first time, the contribution of both substrates was analyzed simultaneously in a single kinetics assay allowing to quantify the contribution of the reversible reaction for kinetics. The protein was crystallized in the spacegroup C222 1 , with unit-cell parameters a = 125.43, b = 148.01, c = 129.76. The structure was solved by molecular replacement and refined at 1.8 Å resolution. In our study, a HEPES molecule was found to interact with HsFH at the C-terminal domain (Domain 3), previously described as involved in allosteric regulation, through a set of interactions that includes Lys 467. HsFH catalytic efficiency is higher when in the presence of HEPES. Mutations at residue 467 have already been implicated in genetic disorders caused by FH deficiency, suggesting that the HEPES-binding site may be important for enzyme kinetics. This study contributes to the understanding of the HsFH structure and how it correlates with mutation, enzymatic deficiency and pathology.
Organizational Affiliation:
Laboratório de Cristalografia de Proteínas, Faculdade de Ciências Farmacêuticas de Ribeirão Preto, Universidade de São Paulo, Ribeirão Preto, Brazil.