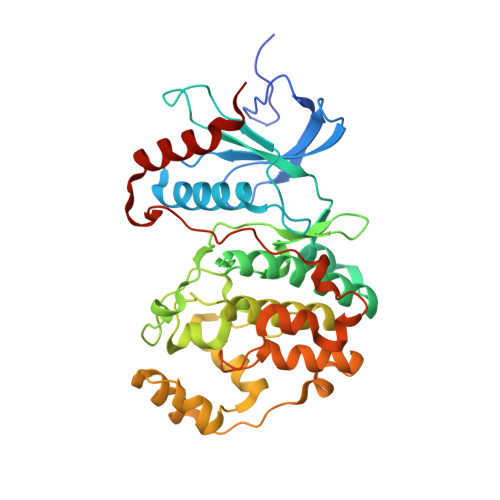
Atomic structure of the MAP kinase ERK2 at 2.3 A resolution.
Zhang, F., Strand, A., Robbins, D., Cobb, M.H., Goldsmith, E.J.(1994) Nature 367: 704-711
- PubMed: 8107865
- DOI: https://doi.org/10.1038/367704a0
- Primary Citation of Related Structures:
5UMO - PubMed Abstract:
The structure of the MAP kinase ERK2, a ubiquitous protein kinase target for regulation by Ras and Raf, has been solved in its unphosphorylated low-activity conformation to a resolution of 2.3 A. The two domains of unphosphorylated ERK2 are farther apart than in the active conformation of cAMP-dependent protein kinase and the peptide-binding site is blocked by tyrosine 185, one of the two residues that are phosphorylated in the active enzyme. Activation of ERK2 is thus likely to involve both global and local conformational changes.
Organizational Affiliation:
Department of Biochemistry, University of Texas Southwestern Medical Center at Dallas.