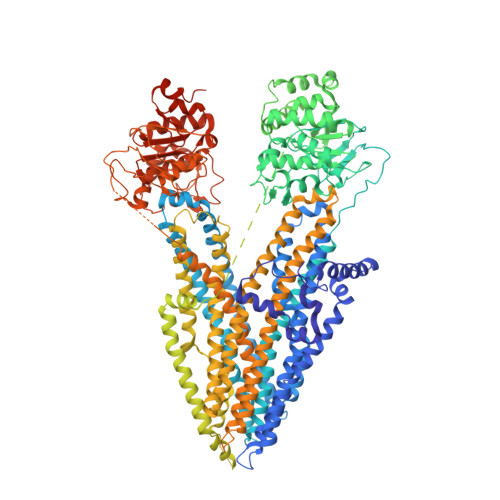
Atomic Structure of the Cystic Fibrosis Transmembrane Conductance Regulator.
Zhang, Z., Chen, J.(2016) Cell 167: 1586-1597.e9
- PubMed: 27912062
- DOI: https://doi.org/10.1016/j.cell.2016.11.014
- Primary Citation of Related Structures:
5UAR - PubMed Abstract:
The cystic fibrosis transmembrane conductance regulator (CFTR) is an anion channel evolved from the ATP-binding cassette (ABC) transporter family. In this study, we determined the structure of zebrafish CFTR in the absence of ATP by electron cryo-microscopy to 3.7 Å resolution. Human and zebrafish CFTR share 55% sequence identity, and 42 of the 46 cystic-fibrosis-causing missense mutational sites are identical. In CFTR, we observe a large anion conduction pathway lined by numerous positively charged residues. A single gate near the extracellular surface closes the channel. The regulatory domain, dephosphorylated, is located in the intracellular opening between the two nucleotide-binding domains (NBDs), preventing NBD dimerization and channel opening. The structure also reveals why many cystic-fibrosis-causing mutations would lead to defects either in folding, ion conduction, or gating and suggests new avenues for therapeutic intervention.
Organizational Affiliation:
The Rockefeller University and Howard Hughes Medical Institute, 1230 York Avenue, New York, NY 10065, USA.