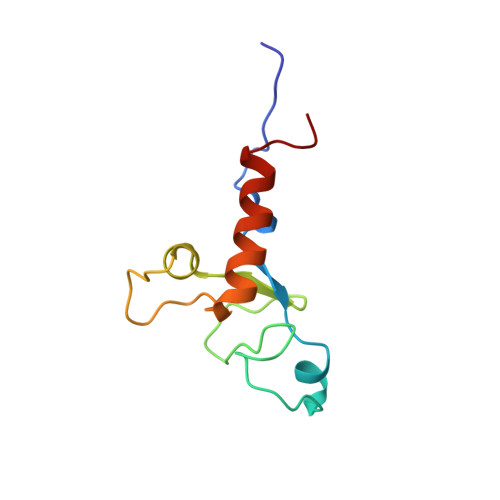
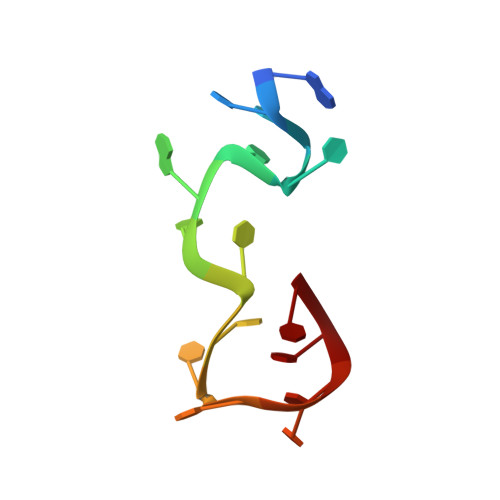Structural Basis for Interaction of the Tandem Zinc Finger Domains of Human Muscleblind with Cognate RNA from Human Cardiac Troponin T.
Park, S., Phukan, P.D., Zeeb, M., Martinez-Yamout, M.A., Dyson, H.J., Wright, P.E.(2017) Biochemistry 56: 4154-4168
- PubMed: 28718627
- DOI: https://doi.org/10.1021/acs.biochem.7b00484
- Primary Citation of Related Structures:
5U6H, 5U6L, 5U9B - PubMed Abstract:
The human muscleblind-like proteins (MBNL) regulate tissue-specific splicing by targeting cardiac troponin T and other pre-mRNAs; aberrant targeting of CUG and CCUG repeat expansions frequently accompanies the neuromuscular disease myotonic dystrophy. We show, using biolayer interferometry (Octet) and NMR spectroscopy, that the zinc finger domains of MBNL isoform 1 (MBNL1) are necessary and sufficient for binding CGCU sequences within the pre-mRNA of human cardiac troponin T. Protein constructs containing zinc fingers 1 and 2 (zf12) and zinc fingers 3 and 4 (zf34) of MBNL1 each fold into a compact globular tandem zinc finger structure that participates in RNA binding. NMR spectra show that the stoichiometry of the interaction between zf12 or zf34 and the CGCU sequence is 1:1, and that the RNA is single-stranded in the complex. The individual zinc fingers within zf12 or zf34 are nonequivalent: the primary RNA binding surface is formed in each pair by the second zinc finger (zf2 or zf4), which interacts with the CGCU RNA sequence. The NMR structure of the complex between zf12 and a 15-base RNA of sequence 95 GUCUCGCUUUUCCCC 109 , containing a single CGCU element, shows the single-stranded RNA wrapped around zf2 and extending to bind to the C-terminal helix. Bases C101, U102, and U103 make well-defined and highly ordered contacts with the protein, whereas neighboring bases are less well-ordered in the complex. Binding of the MBNL zinc fingers to cardiac troponin T pre-mRNA is specific and relatively simple, unlike the complex multiple dimer-trimer stoichiometries postulated in some previous studies.
Organizational Affiliation:
Department of Integrative Structural and Computational Biology and Skaggs Institute of Chemical Biology, The Scripps Research Institute , 10550 North Torrey Pines Road, La Jolla, California 92037, United States.