Crystal Structure of a Full-Length Human Tetraspanin Reveals a Cholesterol-Binding Pocket.
Zimmerman, B., Kelly, B., McMillan, B.J., Seegar, T.C., Dror, R.O., Kruse, A.C., Blacklow, S.C.(2016) Cell 167: 1041-1051.e11
- PubMed: 27881302
- DOI: https://doi.org/10.1016/j.cell.2016.09.056
- Primary Citation of Related Structures:
5TCX - PubMed Abstract:
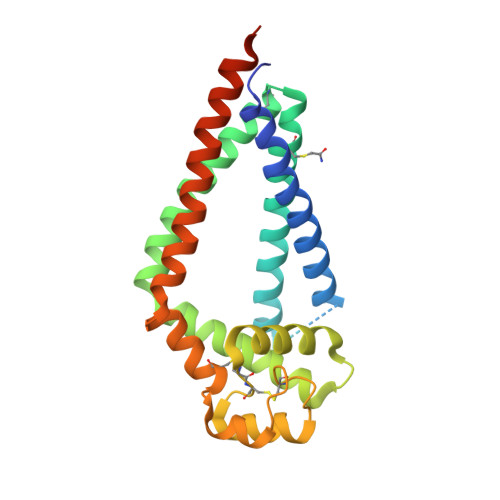
Tetraspanins comprise a diverse family of four-pass transmembrane proteins that play critical roles in the immune, reproductive, genitourinary, and auditory systems. Despite their pervasive roles in human physiology, little is known about the structure of tetraspanins or the molecular mechanisms underlying their various functions. Here, we report the crystal structure of human CD81, a full-length tetraspanin. The transmembrane segments of CD81 pack as two largely separated pairs of helices, capped by the large extracellular loop (EC2) at the outer membrane leaflet. The two pairs of helices converge at the inner leaflet to create an intramembrane pocket with additional electron density corresponding to a bound cholesterol molecule within the cavity. Molecular dynamics simulations identify an additional conformation in which EC2 separates substantially from the transmembrane domain. Cholesterol binding appears to modulate CD81 activity in cells, suggesting a potential mechanism for regulation of tetraspanin function.
Organizational Affiliation:
Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston, MA 02115, USA; Department of Cancer Biology, Dana Farber Cancer Institute, Boston, MA 02215, USA.