Direct binding of CEP85 to STIL ensures robust PLK4 activation and efficient centriole assembly.
Liu, Y., Gupta, G.D., Barnabas, D.D., Agircan, F.G., Mehmood, S., Wu, D., Coyaud, E., Johnson, C.M., McLaughlin, S.H., Andreeva, A., Freund, S.M.V., Robinson, C.V., Cheung, S.W.T., Raught, B., Pelletier, L., van Breugel, M.(2018) Nat Commun 9: 1731-1731
- PubMed: 29712910
- DOI: https://doi.org/10.1038/s41467-018-04122-x
- Primary Citation of Related Structures:
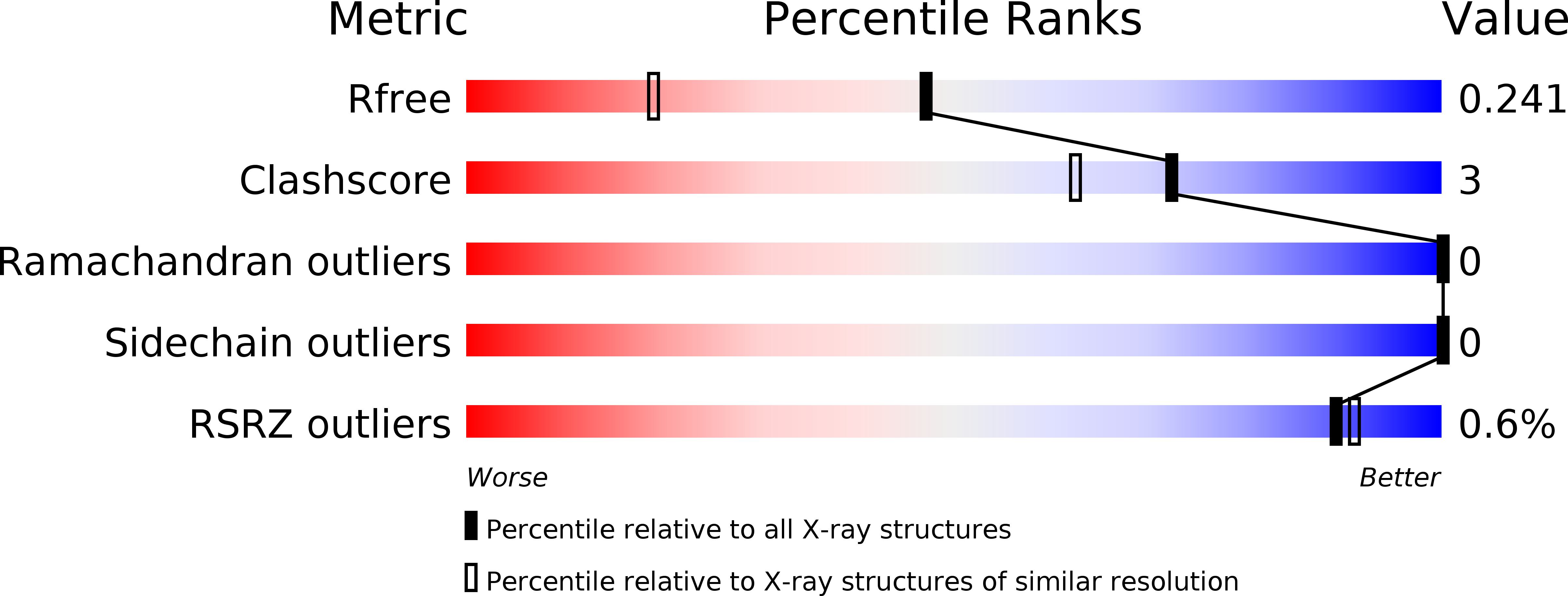
5OI7, 5OI9, 5OID - PubMed Abstract:
Centrosomes are required for faithful chromosome segregation during mitosis. They are composed of a centriole pair that recruits and organizes the microtubule-nucleating pericentriolar material. Centriole duplication is tightly controlled in vivo and aberrations in this process are associated with several human diseases, including cancer and microcephaly. Although factors essential for centriole assembly, such as STIL and PLK4, have been identified, the underlying molecular mechanisms that drive this process are incompletely understood. Combining protein proximity mapping with high-resolution structural methods, we identify CEP85 as a centriole duplication factor that directly interacts with STIL through a highly conserved interaction interface involving a previously uncharacterised domain of STIL. Structure-guided mutational analyses in vivo demonstrate that this interaction is essential for efficient centriolar targeting of STIL, PLK4 activation and faithful daughter centriole assembly. Taken together, our results illuminate a molecular mechanism underpinning the spatiotemporal regulation of the early stages of centriole duplication.
Organizational Affiliation:
Lunenfeld-Tanenbaum Research Institute, Sinai Health System, 600 University Avenue, Toronto, ON M5G 1X5, Canada.