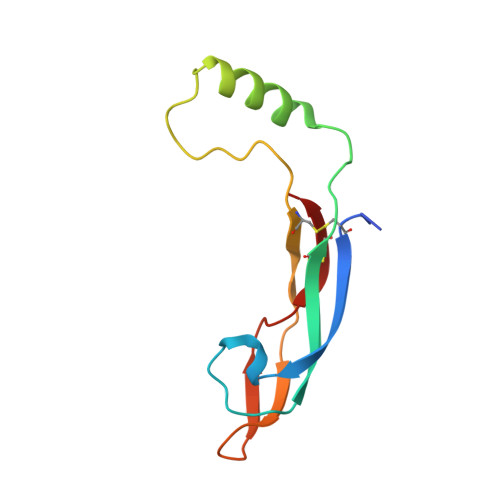
Structure and biophysical characterisation of the human complex - a role for heparan sulfate
Sandmark, J., Dahl, G., Oster, L., Xu, B., Johansson, P., Akerud, T., Aagaard, A., Davidsson, P., Sorhede-Winzell, M., Rainey, G.J., Roth, R.G.To be published.