Immunoglobulin domain interface exchange as a platform technology for the generation of Fc heterodimers and bispecific antibodies.
Skegro, D., Stutz, C., Ollier, R., Svensson, E., Wassmann, P., Bourquin, F., Monney, T., Gn, S., Blein, S.(2017) J Biol Chem 292: 9745-9759
- PubMed: 28450393
- DOI: https://doi.org/10.1074/jbc.M117.782433
- Primary Citation of Related Structures:
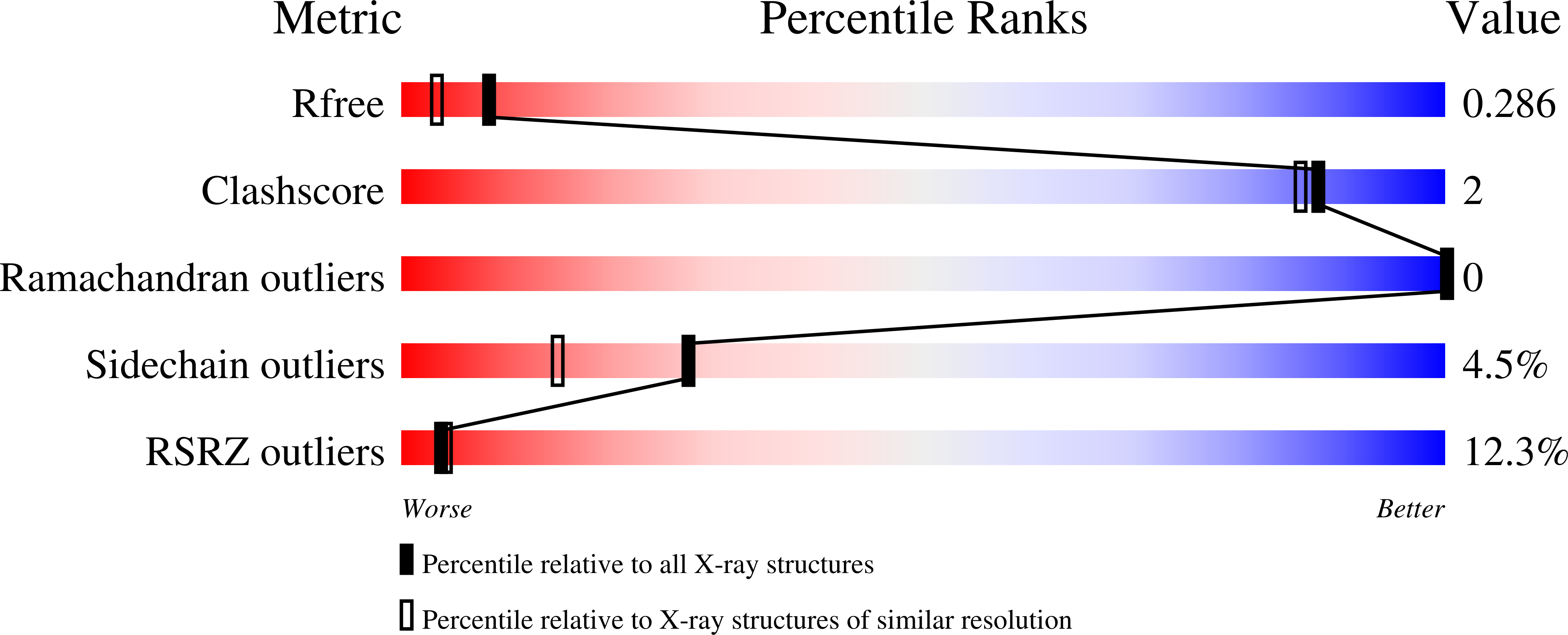
5M3V - PubMed Abstract:
Bispecific antibodies (bsAbs) are of significant importance to the development of novel antibody-based therapies, and heavy chain (Hc) heterodimers represent a major class of bispecific drug candidates. Current technologies for the generation of Hc heterodimers are suboptimal and often suffer from contamination by homodimers posing purification challenges. Here, we introduce a new technology based on biomimicry wherein the protein-protein interfaces of two different immunoglobulin (Ig) constant domain pairs are exchanged in part or fully to design new heterodimeric domains. The method can be applied across Igs to design Fc heterodimers and bsAbs. We investigated interfaces from human IgA CH3, IgD CH3, IgG1 CH3, IgM CH4, T-cell receptor (TCR) α/β, and TCR γ/δ constant domain pairs, and we found that they successfully drive human IgG1 CH3 or IgM CH4 heterodimerization to levels similar to or above those of reference methods. A comprehensive interface exchange between the TCR α/β constant domain pair and the IgG1 CH3 homodimer was evidenced by X-ray crystallography and used to engineer examples of bsAbs for cancer therapy. Parental antibody pairs were rapidly reformatted into scalable bsAbs that were free of homodimer traces by combining interface exchange, asymmetric Protein A binding, and the scFv × Fab format. In summary, we successfully built several new CH3- or CH4-based heterodimers that may prove useful for designing new bsAb-based therapeutics, and we anticipate that our approach could be broadly implemented across the Ig constant domain family. To our knowledge, CH4-based heterodimers have not been previously reported.
Organizational Affiliation:
From the Department of Antibody Engineering, Biologics Research, Glenmark Pharmaceuticals S.A., Chemin de la Combeta 5, 2300 La Chaux-de-Fonds, Switzerland and.