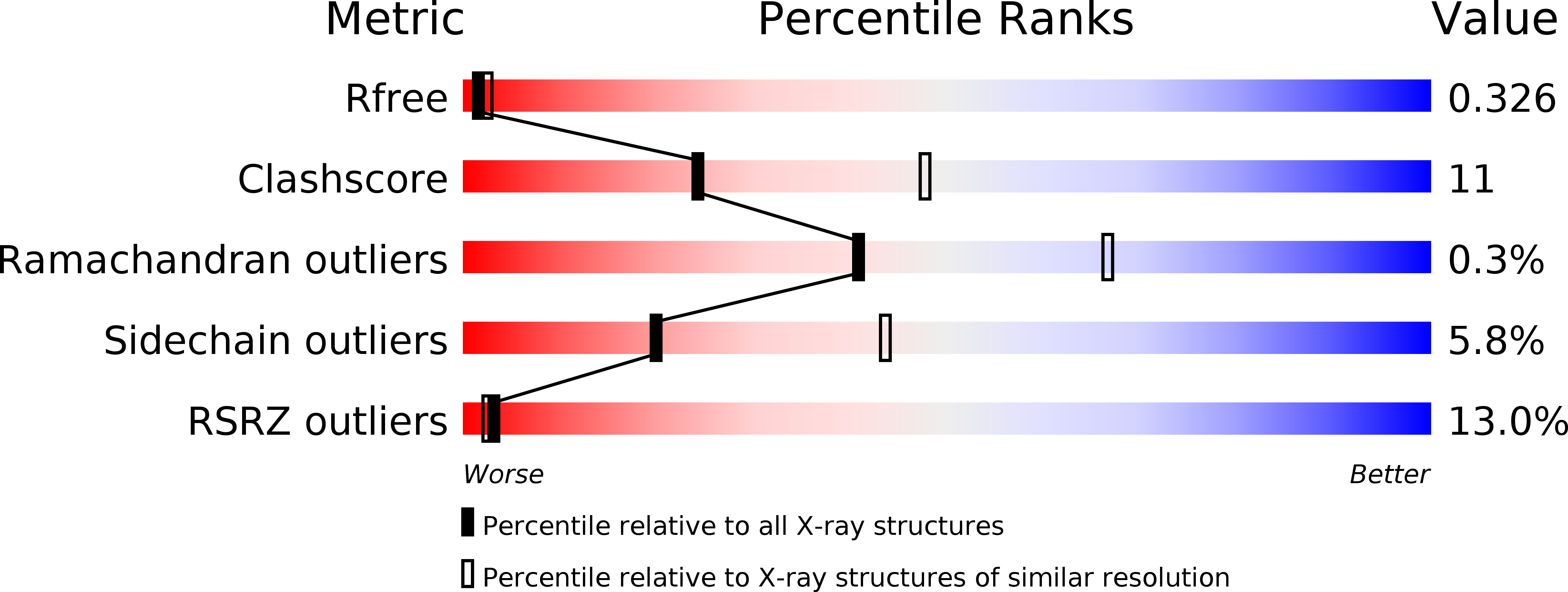
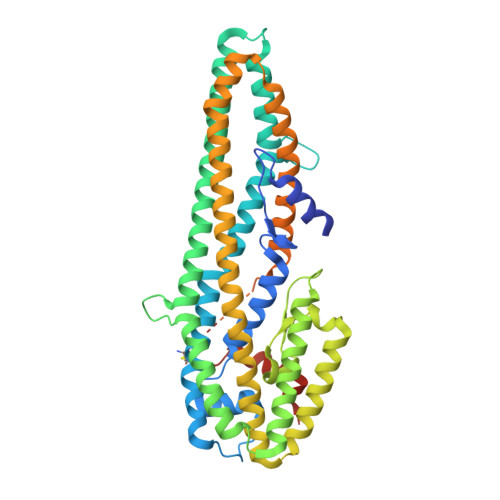
The pesticidal Cry6Aa toxin from Bacillus thuringiensis is structurally similar to HlyE-family alpha pore-forming toxins.
Dementiev, A., Board, J., Sitaram, A., Hey, T., Kelker, M.S., Xu, X., Hu, Y., Vidal-Quist, C., Chikwana, V., Griffin, S., McCaskill, D., Wang, N.X., Hung, S.C., Chan, M.K., Lee, M.M., Hughes, J., Wegener, A., Aroian, R.V., Narva, K.E., Berry, C.(2016) BMC Biol 14: 71-71
- PubMed: 27576487
- DOI: https://doi.org/10.1186/s12915-016-0295-9
- Primary Citation of Related Structures:
5KUC, 5KUD - PubMed Abstract:
The Cry6 family of proteins from Bacillus thuringiensis represents a group of powerful toxins with great potential for use in the control of coleopteran insects and of nematode parasites of importance to agriculture. These proteins are unrelated to other insecticidal toxins at the level of their primary sequences and the structure and function of these proteins has been poorly studied to date. This has inhibited our understanding of these toxins and their mode of action, along with our ability to manipulate the proteins to alter their activity to our advantage. To increase our understanding of their mode of action and to facilitate further development of these proteins we have determined the structure of Cry6Aa in protoxin and trypsin-activated forms and demonstrated a pore-forming mechanism of action. The two forms of the toxin were resolved to 2.7 Å and 2.0 Å respectively and showed very similar structures. Cry6Aa shows structural homology to a known class of pore-forming toxins including hemolysin E from Escherichia coli and two Bacillus cereus proteins: the hemolytic toxin HblB and the NheA component of the non-hemolytic toxin (pfam05791). Cry6Aa also shows atypical features compared to other members of this family, including internal repeat sequences and small loop regions within major alpha helices. Trypsin processing was found to result in the loss of some internal sequences while the C-terminal region remains disulfide-linked to the main core of the toxin. Based on the structural similarity of Cry6Aa to other toxins, the mechanism of action of the toxin was probed and its ability to form pores in vivo in Caenorhabditis elegans was demonstrated. A non-toxic mutant was also produced, consistent with the proposed pore-forming mode of action. Cry6 proteins are members of the alpha helical pore-forming toxins - a structural class not previously recognized among the Cry toxins of B. thuringiensis and representing a new paradigm for nematocidal and insecticidal proteins. Elucidation of both the structure and the pore-forming mechanism of action of Cry6Aa now opens the way to more detailed analysis of toxin specificity and the development of new toxin variants with novel activities.
Organizational Affiliation:
Shamrock Structures LLC, Woodridge, IL, USA.