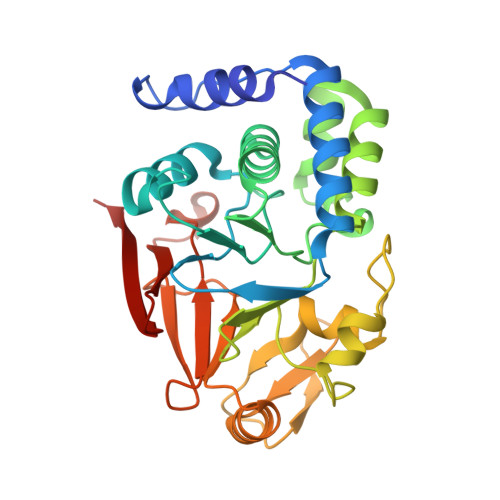
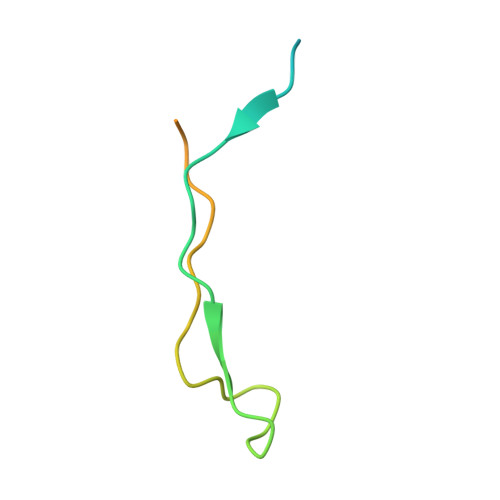
The Ki-67 and RepoMan mitotic phosphatases assemble via an identical, yet novel mechanism.
Kumar, G.S., Gokhan, E., De Munter, S., Bollen, M., Vagnarelli, P., Peti, W., Page, R.(2016) Elife 5
- PubMed: 27572260
- DOI: https://doi.org/10.7554/eLife.16539
- Primary Citation of Related Structures:
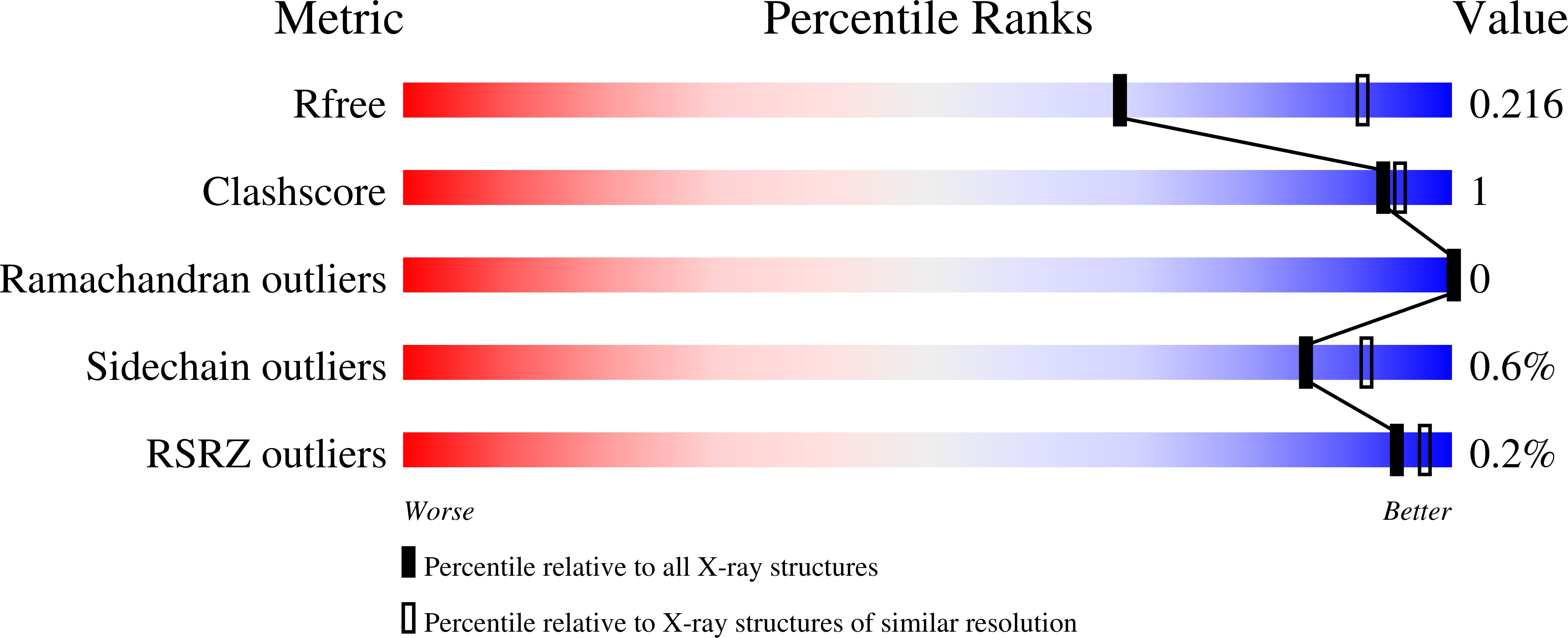
5INB, 5IOH, 5J28 - PubMed Abstract:
Ki-67 and RepoMan have key roles during mitotic exit. Previously, we showed that Ki-67 organizes the mitotic chromosome periphery and recruits protein phosphatase 1 (PP1) to chromatin at anaphase onset, in a similar manner as RepoMan (Booth et al., 2014). Here we show how Ki-67 and RepoMan form mitotic exit phosphatases by recruiting PP1, how they distinguish between distinct PP1 isoforms and how the assembly of these two holoenzymes are dynamically regulated by Aurora B kinase during mitosis. Unexpectedly, our data also reveal that Ki-67 and RepoMan bind PP1 using an identical, yet novel mechanism, interacting with a PP1 pocket that is engaged only by these two PP1 regulators. These findings not only show how two distinct mitotic exit phosphatases are recruited to their substrates, but also provide immediate opportunities for the design of novel cancer therapeutics that selectively target the Ki-67:PP1 and RepoMan:PP1 holoenzymes.
Organizational Affiliation:
Department of Molecular Pharmacology, Physiology and Biotechnology, Brown University, Providence, United States.