Novel hyperoxidation resistance motifs in 2-Cys peroxiredoxins.
Bolduc, J.A., Nelson, K.J., Haynes, A.C., Lee, J., Reisz, J.A., Graff, A.H., Clodfelter, J.E., Parsonage, D., Poole, L.B., Furdui, C.M., Lowther, W.T.(2018) J Biol Chem 293: 11901-11912
- PubMed: 29884768
- DOI: https://doi.org/10.1074/jbc.RA117.001690
- Primary Citation of Related Structures:
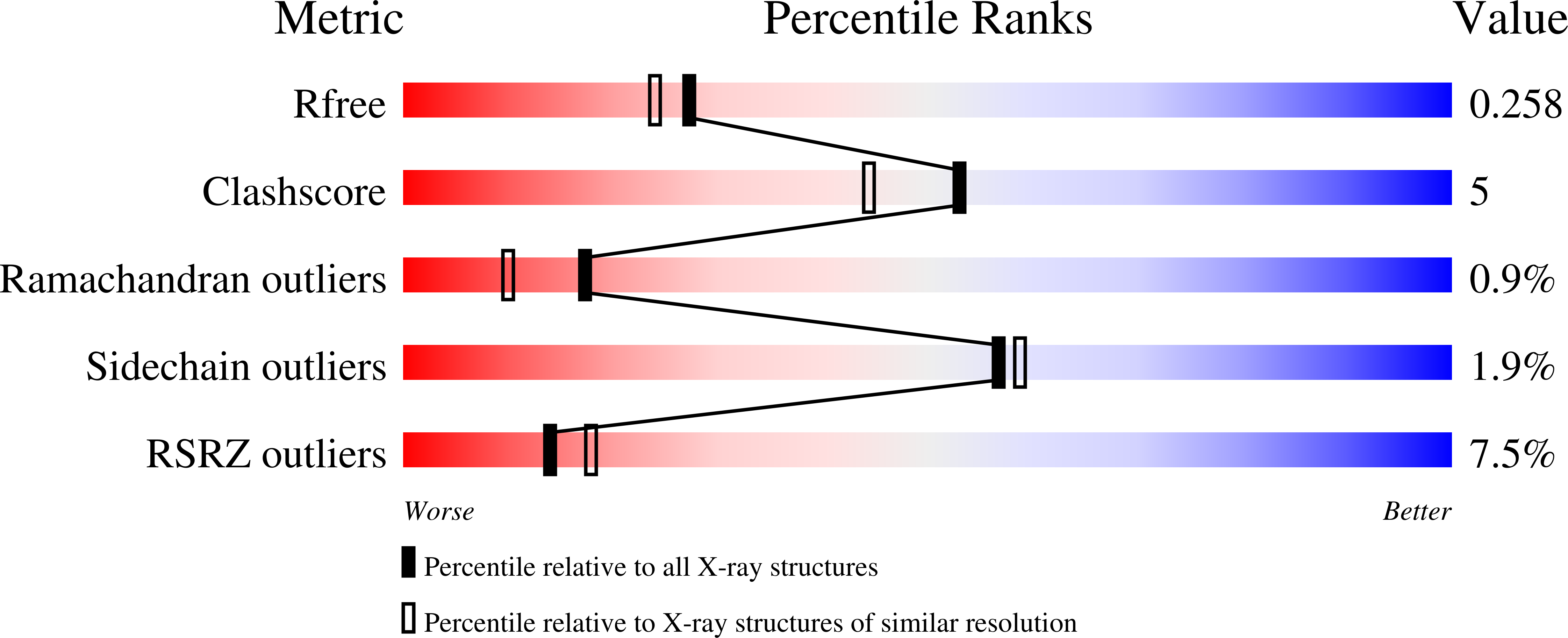
5IJT - PubMed Abstract:
2-Cys peroxiredoxins (Prxs) modulate hydrogen peroxide (H 2 O 2 )-mediated cell signaling. At high H 2 O 2 levels, eukaryotic Prxs can be inactivated by hyperoxidation and are classified as sensitive Prxs. In contrast, prokaryotic Prxs are categorized as being resistant to hyperoxidation and lack the GGLG and C-terminal YF motifs present in the sensitive Prxs. Additional molecular determinants that account for the subtle differences in the susceptibility to hyperoxidation remain to be identified. A comparison of a new, 2.15-Å-resolution crystal structure of Prx2 in the oxidized, disulfide-bonded state with the hyperoxidized structure of Prx2 and Prx1 in complex with sulfiredoxin revealed three structural regions that rearrange during catalysis. With these regions in hand, focused sequence analyses were performed comparing sensitive and resistant Prx groups. From this combinatorial approach, we discovered two novel hyperoxidation resistance motifs, motifs A and B, which were validated using mutagenesis of sensitive human Prxs and resistant Salmonella enterica serovar Typhimurium AhpC. Introduction and removal of these motifs, respectively, resulted in drastic changes in the sensitivity to hyperoxidation with Prx1 becoming 100-fold more resistant to hyperoxidation and AhpC becoming 800-fold more sensitive to hyperoxidation. The increased sensitivity of the latter AhpC variant was also confirmed in vivo These results support the function of motifs A and B as primary drivers for tuning the sensitivity of Prxs to different levels of H 2 O 2 , thus enabling the initiation of variable signaling or antioxidant responses in cells.
Organizational Affiliation:
From the Center for Structural Biology, Department of Biochemistry.