Structural Insight into the Critical Role of the N-Terminal Region in the Catalytic Activity of Dual-Specificity Phosphatase 26
Won, E.Y., Lee, S.O., Lee, D.H., Lee, D., Bae, K.H., Lee, S.C., Kim, S.J., Chi, S.W.(2016) PLoS One 11: e0162115-e0162115
- PubMed: 27583453
- DOI: https://doi.org/10.1371/journal.pone.0162115
- Primary Citation of Related Structures:
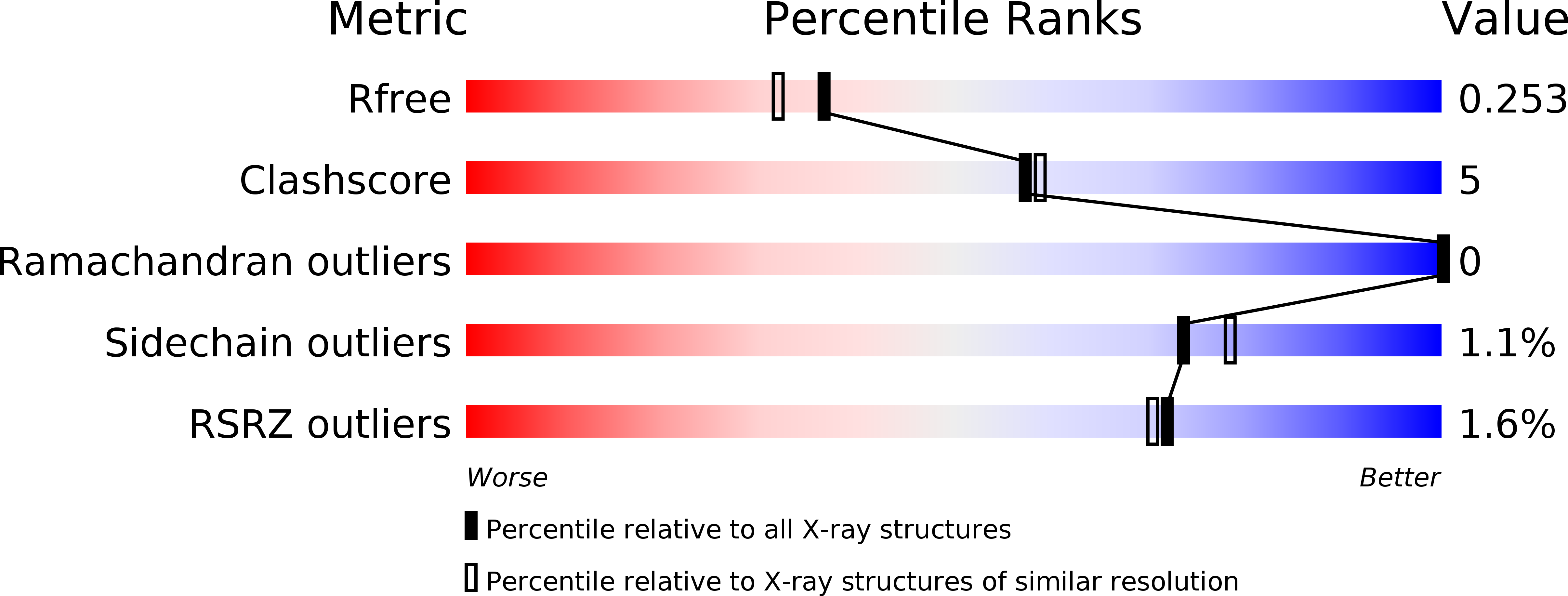
5GTJ - PubMed Abstract:
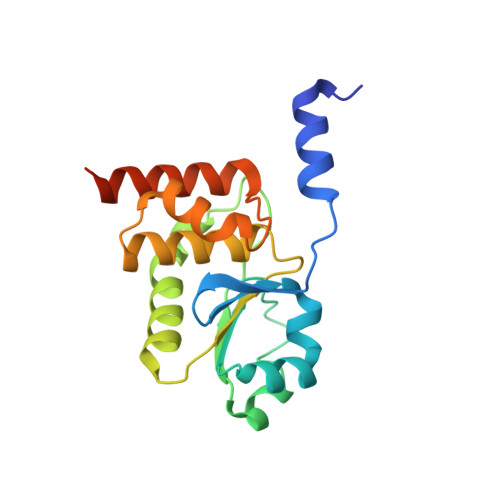
Human dual-specificity phosphatase 26 (DUSP26) is a novel target for anticancer therapy because its dephosphorylation of the p53 tumor suppressor regulates the apoptosis of cancer cells. DUSP26 inhibition results in neuroblastoma cell cytotoxicity through p53-mediated apoptosis. Despite the previous structural studies of DUSP26 catalytic domain (residues 61-211, DUSP26-C), the high-resolution structure of its catalytically active form has not been resolved. In this study, we determined the crystal structure of a catalytically active form of DUSP26 (residues 39-211, DUSP26-N) with an additional N-terminal region at 2.0 Å resolution. Unlike the C-terminal domain-swapped dimeric structure of DUSP26-C, the DUSP26-N (C152S) monomer adopts a fold-back conformation of the C-terminal α8-helix and has an additional α1-helix in the N-terminal region. Consistent with the canonically active conformation of its protein tyrosine phosphate-binding loop (PTP loop) observed in the structure, the phosphatase assay results demonstrated that DUSP26-N has significantly higher catalytic activity than DUSP26-C. Furthermore, size exclusion chromatography-multiangle laser scattering (SEC-MALS) measurements showed that DUSP26-N (C152S) exists as a monomer in solution. Notably, the crystal structure of DUSP26-N (C152S) revealed that the N-terminal region of DUSP26-N (C152S) serves a scaffolding role by positioning the surrounding α7-α8 loop for interaction with the PTP-loop through formation of an extensive hydrogen bond network, which seems to be critical in making the PTP-loop conformation competent for phosphatase activity. Our study provides the first high-resolution structure of a catalytically active form of DUSP26, which will contribute to the structure-based rational design of novel DUSP26-targeting anticancer therapeutics.
Organizational Affiliation:
Disease Target Structure Research Center, Korea Research Institute of Bioscience and Biotechnology, Daejeon, Republic of Korea.