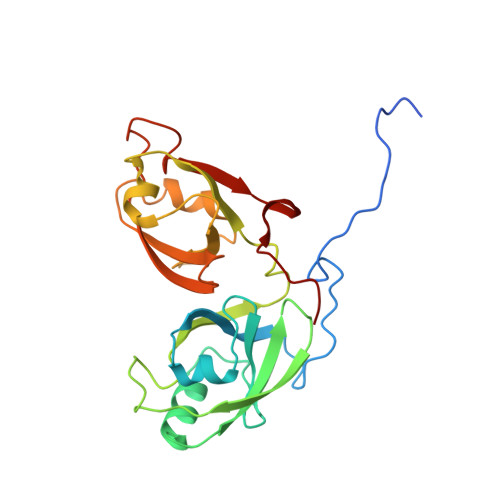
Genetic, structural, and chemical insights into the dual function of GRASP55 in germ cell Golgi remodeling and JAM-C polarized localization during spermatogenesis
Cartier-Michaud, A., Bailly, A.L., Betzi, S., Shi, X., Lissitzky, J.C., Zarubica, A., Serge, A., Roche, P., Lugari, A., Hamon, V., Bardin, F., Derviaux, C., Lembo, F., Audebert, S., Marchetto, S., Durand, B., Borg, J.P., Shi, N., Morelli, X., Aurrand-Lions, M.(2017) PLoS Genet 13: e1006803-e1006803
- PubMed: 28617811
- DOI: https://doi.org/10.1371/journal.pgen.1006803
- Primary Citation of Related Structures:
5GMI, 5GMJ - PubMed Abstract:
Spermatogenesis is a dynamic process that is regulated by adhesive interactions between germ and Sertoli cells. Germ cells express the Junctional Adhesion Molecule-C (JAM-C, encoded by Jam3), which localizes to germ/Sertoli cell contacts. JAM-C is involved in germ cell polarity and acrosome formation. Using a proteomic approach, we demonstrated that JAM-C interacted with the Golgi reassembly stacking protein of 55 kDa (GRASP55, encoded by Gorasp2) in developing germ cells. Generation and study of Gorasp2-/- mice revealed that knock-out mice suffered from spermatogenesis defects. Acrosome formation and polarized localization of JAM-C in spermatids were altered in Gorasp2-/- mice. In addition, Golgi morphology of spermatocytes was disturbed in Gorasp2-/- mice. Crystal structures of GRASP55 in complex with JAM-C or JAM-B revealed that GRASP55 interacted via PDZ-mediated interactions with JAMs and induced a conformational change in GRASP55 with respect of its free conformation. An in silico pharmacophore approach identified a chemical compound called Graspin that inhibited PDZ-mediated interactions of GRASP55 with JAMs. Treatment of mice with Graspin hampered the polarized localization of JAM-C in spermatids, induced the premature release of spermatids and affected the Golgi morphology of meiotic spermatocytes.
Organizational Affiliation:
Aix Marseille Univ, CNRS, INSERM, Institut Paoli-Calmettes, CRCM, Marseille, France.