A novel cysteine carbamoyl-switch is responsible for the inhibition of formamidase, a nitrilase superfamily member.
Martinez-Rodriguez, S., Conejero-Muriel, M., Gavira, J.A.(2019) Arch Biochem Biophys 662: 151-159
- PubMed: 30528776
- DOI: https://doi.org/10.1016/j.abb.2018.12.008
- Primary Citation of Related Structures:
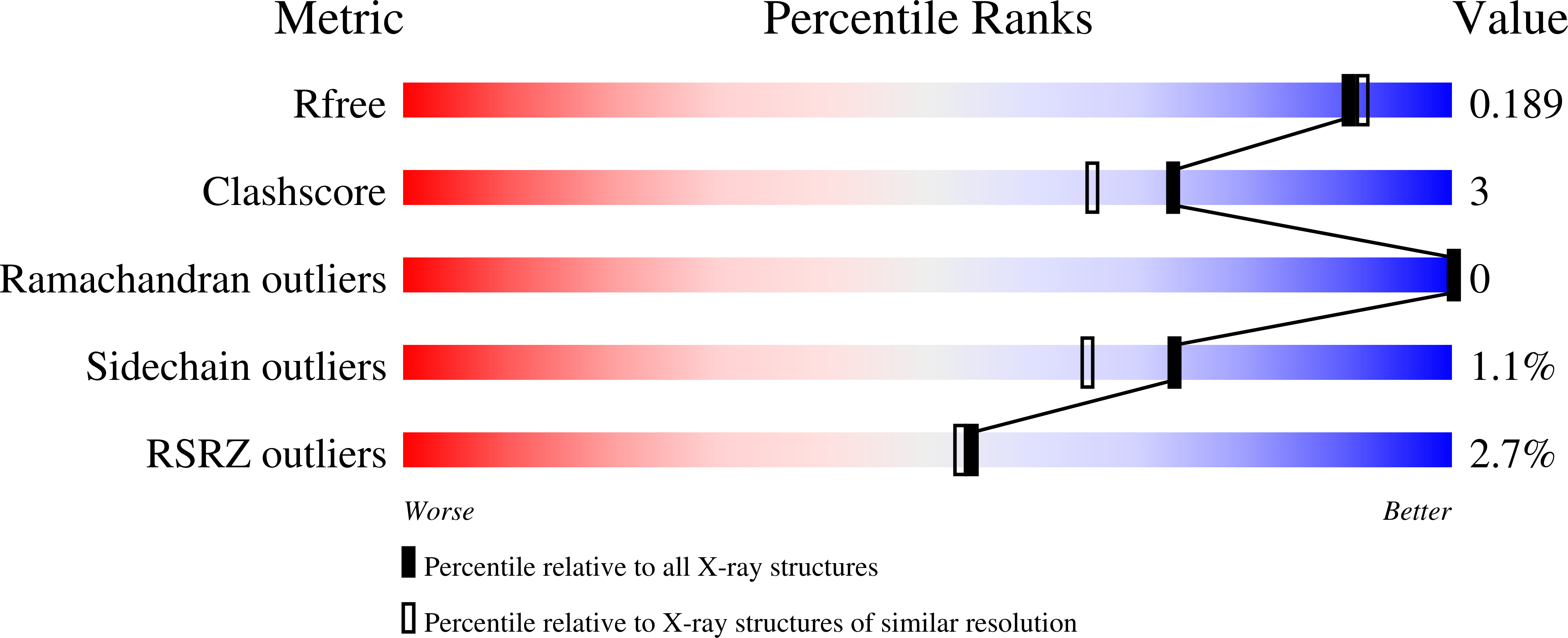
5G3O, 5G3P - PubMed Abstract:
Formamidases (EC 3.5.1.49) and amidases (EC 3.5.1.4) are paralogous cysteine-dependent enzymes which catalyze the conversion of amide substrates to ammonia and the corresponding carboxylic acid. Both enzymes have been suggested as an alternative pathway for ammonia production during urea shortage. Urea was proved key in the transcriptional regulation of formamidases/amidases, connecting urea level to amide metabolism. In addition, different amidases have also been shown to be inhibited by urea, pointing to urea-regulation at the enzymatic level. Although amidases have been widely studied due to its biotechnological application in the hydrolysis of aliphatic amides, up to date, only two formamidases have been extensively characterized, belonging to Helicobacter pylori (HpyAmiF) and Bacillus cereus (BceAmiF). In this work, we report the first structure of an acyl-intermediate of BceAmiF. We also report the inhibition of BceAmiF by urea, together with mass spectrometry studies confirming the S-carbamoylation of BceAmiF after urea treatment. X-ray studies of urea-soaked BceAmiF crystals showed short- and long-range rearrangements affecting oligomerization interfaces. Since cysteine-based switches are known to occur in the regulation of different metabolic and signaling pathways, our results suggest a novel S-carbamoylation-switch for the regulation of BceAmiF. This finding could relate to previous observations of unexplained modifications in the catalytic cysteine of different nitrilase superfamily members and therefore extending this regulation mechanism to the whole nitrilase superfamily.
Organizational Affiliation:
Departamento de Bioquímica y Biología Molecular III e Inmunología, Universidad de Granada (Campus de Melilla), 52071, Melilla, Spain; Laboratorio de Estudios Cristalográficos, CSIC-UGR, 18100, Granada, Spain. Electronic address: sergio@ugr.es.