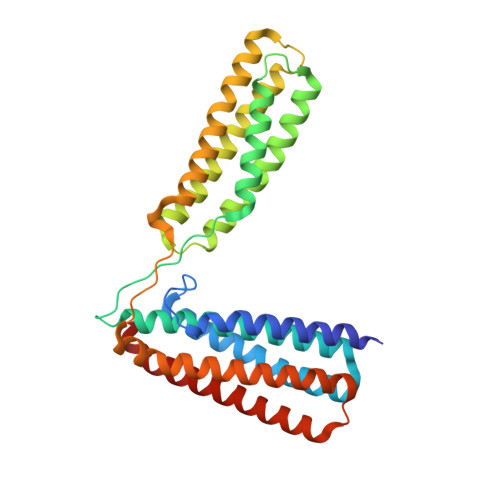
Ld Motif Recognition by Talin: Structure of the Talin-Dlc1 Complex.
Zacharchenko, T., Qian, X., Goult, B.T., Jethwa, D., Almeida, T.B., Ballestrem, C., Critchley, D.R., Lowy, D.R., Barsukov, I.L.(2016) Structure 24: 1130
- PubMed: 27265849
- DOI: https://doi.org/10.1016/j.str.2016.04.016
- Primary Citation of Related Structures:
5FZT - PubMed Abstract:
Cell migration requires coordination between integrin-mediated cell adhesion to the extracellular matrix and force applied to adhesion sites. Talin plays a key role in coupling integrin receptors to the actomyosin contractile machinery, while deleted in liver cancer 1 (DLC1) is a Rho GAP that binds talin and regulates Rho, and therefore actomyosin contractility. We show that the LD motif of DLC1 forms a helix that binds to the four-helix bundle of the talin R8 domain in a canonical triple-helix arrangement. We demonstrate that the same R8 surface interacts with the paxillin LD1 and LD2 motifs. We identify key charged residues that stabilize the R8 interactions with LD motifs and demonstrate their importance in vitro and in cells. Our results suggest a network of competitive interactions in adhesion complexes that involve LD motifs, and identify mutations that can be used to analyze the biological roles of specific protein-protein interactions in cell migration.
Organizational Affiliation:
Institute of Integrative Biology, University of Liverpool, BioSciences Building, Crown Street, Liverpool L69 7ZB, UK.