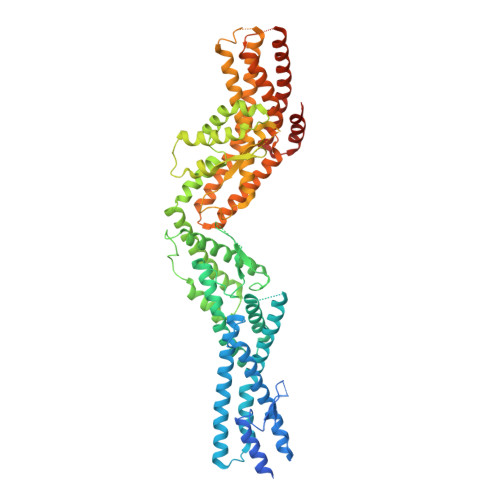
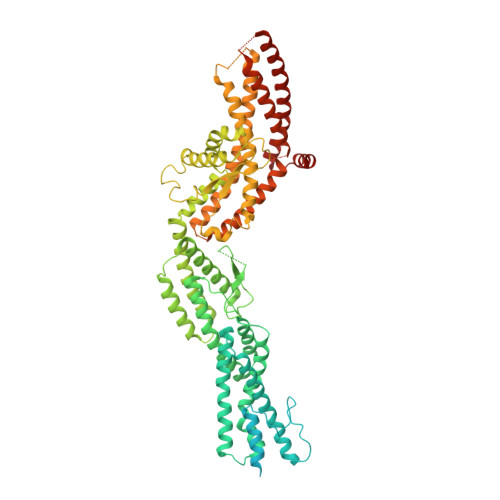
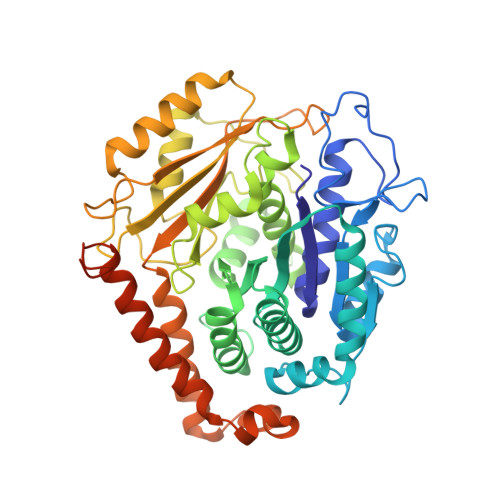
Structure of Gamma-Tubulin Small Complex Based on a Cryo-Em Map, Chemical Cross-Links, and a Remotely Related Structure.
Greenberg, C.H., Kollman, J., Zelter, A., Johnson, R., Maccoss, M.J., Davis, T.N., Agard, D.A., Sali, A.(2016) J Struct Biol 194: 303
- PubMed: 26968363
- DOI: https://doi.org/10.1016/j.jsb.2016.03.006
- Primary Citation of Related Structures:
5FLZ, 5FM1 - PubMed Abstract:
Modeling protein complex structures based on distantly related homologues can be challenging due to poor sequence and structure conservation. Therefore, utilizing even low-resolution experimental data can significantly increase model precision and accuracy. Here, we present models of the two key functional states of the yeast γ-tubulin small complex (γTuSC): one for the low-activity "open" state and another for the higher-activity "closed" state. Both models were computed based on remotely related template structures and cryo-EM density maps at 6.9Å and 8.0Å resolution, respectively. For each state, extensive sampling of alignments and conformations was guided by the fit to the corresponding cryo-EM density map. The resulting good-scoring models formed a tightly clustered ensemble of conformations in most regions. We found significant structural differences between the two states, primarily in the γ-tubulin subunit regions where the microtubule binds. We also report a set of chemical cross-links that were found to be consistent with equilibrium between the open and closed states. The protocols developed here have been incorporated into our open-source Integrative Modeling Platform (IMP) software package (http://integrativemodeling.org), and can therefore be applied to many other systems.
- Department of Bioengineering and Therapeutic Sciences, Department of Pharmaceutical Chemistry, California Institute for Quantitative Biosciences, University of California at San Francisco, San Francisco, CA, USA. Electronic address: cgreen@salilab.org.
Organizational Affiliation: