Identification of N-(4-((1R,3S,5S)-3-Amino-5-methylcyclohexyl)pyridin-3-yl)-6-(2,6-difluorophenyl)-5-fluoropicolinamide (PIM447), a Potent and Selective Proviral Insertion Site of Moloney Murine Leukemia (PIM) 1, 2, and 3 Kinase Inhibitor in Clinical Trials for Hematological Malignancies.
Burger, M.T., Nishiguchi, G., Han, W., Lan, J., Simmons, R., Atallah, G., Ding, Y., Tamez, V., Zhang, Y., Mathur, M., Muller, K., Bellamacina, C., Lindvall, M.K., Zang, R., Huh, K., Feucht, P., Zavorotinskaya, T., Dai, Y., Basham, S., Chan, J., Ginn, E., Aycinena, A., Holash, J., Castillo, J., Langowski, J.L., Wang, Y., Chen, M.Y., Lambert, A., Fritsch, C., Kauffmann, A., Pfister, E., Vanasse, K.G., Garcia, P.D.(2015) J Med Chem 58: 8373-8386
- PubMed: 26505898
- DOI: https://doi.org/10.1021/acs.jmedchem.5b01275
- Primary Citation of Related Structures:
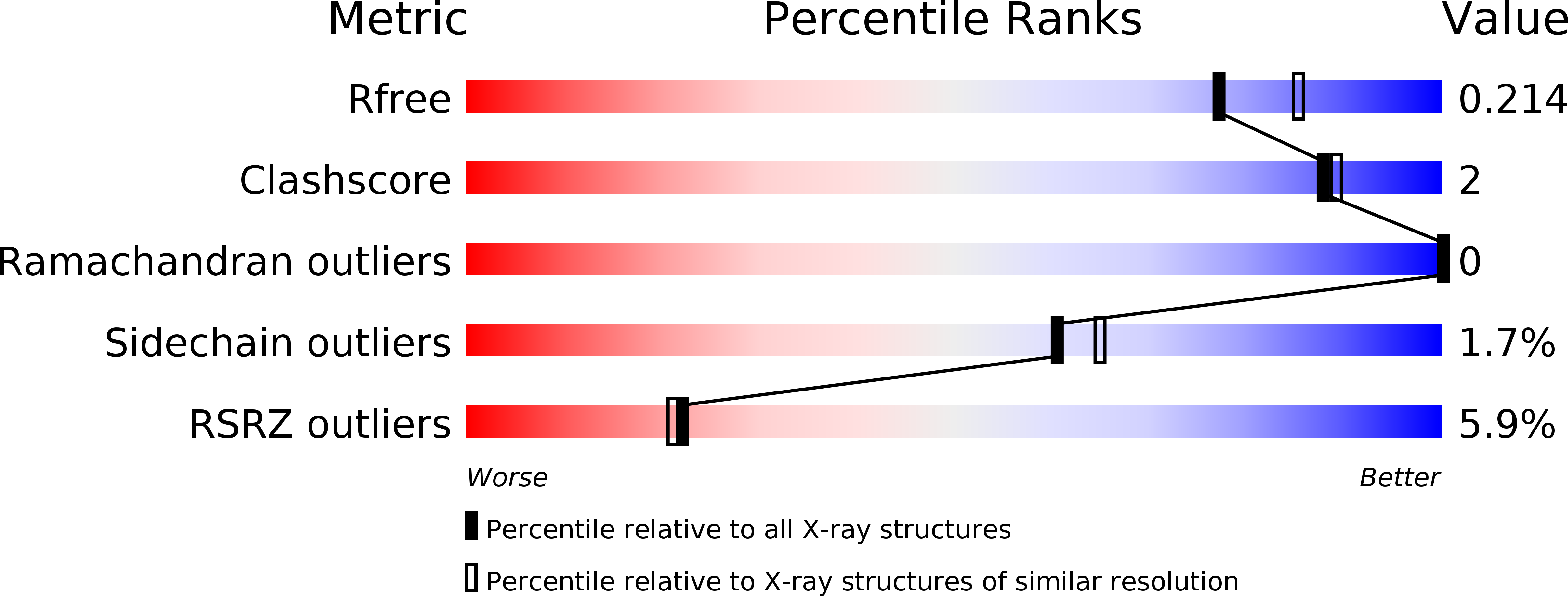
5DWR - PubMed Abstract:
Pan proviral insertion site of Moloney murine leukemia (PIM) 1, 2, and 3 kinase inhibitors have recently begun to be tested in humans to assess whether pan PIM kinase inhibition may provide benefit to cancer patients. Herein, the synthesis, in vitro activity, in vivo activity in an acute myeloid leukemia xenograft model, and preclinical profile of the potent and selective pan PIM kinase inhibitor compound 8 (PIM447) are described. Starting from the reported aminopiperidyl pan PIM kinase inhibitor compound 3, a strategy to improve the microsomal stability was pursued resulting in the identification of potent aminocyclohexyl pan PIM inhibitors with high metabolic stability. From this aminocyclohexyl series, compound 8 entered the clinic in 2012 in multiple myeloma patients and is currently in several phase 1 trials of cancer patients with hematological malignancies.
Organizational Affiliation:
Oncology Research, Novartis Institutes for Biomedical Research , CH-4056, Basel, Switzerland.