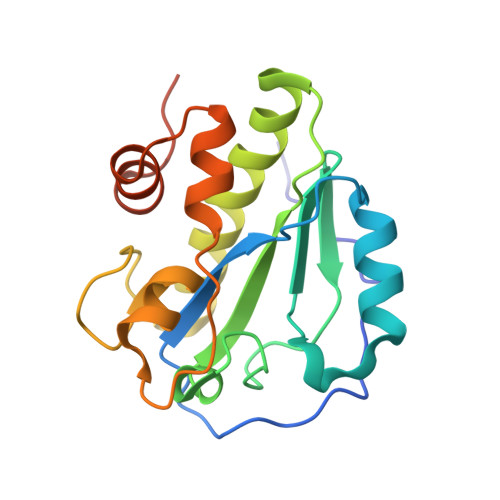
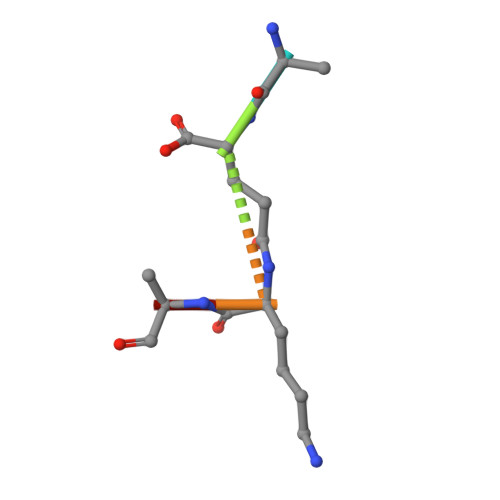
The crystal structure of the major pneumococcal autolysin LytA in complex with a large peptidoglycan fragment reveals the pivotal role of glycans for lytic activity.
Sandalova, T., Lee, M., Henriques-Normark, B., Hesek, D., Mobashery, S., Mellroth, P., Achour, A.(2016) Mol Microbiol 101: 954-967
- PubMed: 27273793
- DOI: https://doi.org/10.1111/mmi.13435
- Primary Citation of Related Structures:
5CTV - PubMed Abstract:
The pneumococcal autolysin LytA is a key virulence factor involved in several important functions including DNA competence, immune evasion and biofilm formation. Here, we present the 1.05 Å crystal structure of the catalytic domain of LytA in complex with a synthetic cell-wall-based peptidoglycan (PG) ligand that occupies the entire Y-shaped substrate-binding crevice. As many as twenty-one amino-acid residues are engaged in ligand interactions with a majority of these interactions directed towards the glycan strand. All saccharides are intimately bound through hydrogen bond, van der Waals and CH-π interactions. Importantly, the structure of LytA is not altered upon ligand binding, whereas the bound ligand assumes a different conformation compared to the unbound NMR-based solution structure of the same PG-fragment. Mutational study reveals that several non-catalytic glycan-interacting residues, structurally conserved in other amidases from Gram-positive Firmicutes, are pivotal for enzymatic activity. The three-dimensional structure of the LytA/PG complex provides a novel structural basis for ligand restriction by the pneumococcal autolysin, revealing for the first time an importance of the multivalent binding to PG saccharides.
Organizational Affiliation:
Science for Life Laboratory, Department of Medicine Solna, Karolinska Institutet, and Department of Infectious Diseases, Karolinska University Hospital, Solna, Stockholm, SE, 17176, Sweden.