Concomitant Binding of Afadin to Lgn and F-Actin Directs Planar Spindle Orientation.
Carminati, M., Gallini, S., Pirovano, L., Alfieri, A., Bisi, S., Mapelli, M.(2016) Nat Struct Mol Biol 23: 155
- PubMed: 26751642
- DOI: https://doi.org/10.1038/nsmb.3152
- Primary Citation of Related Structures:
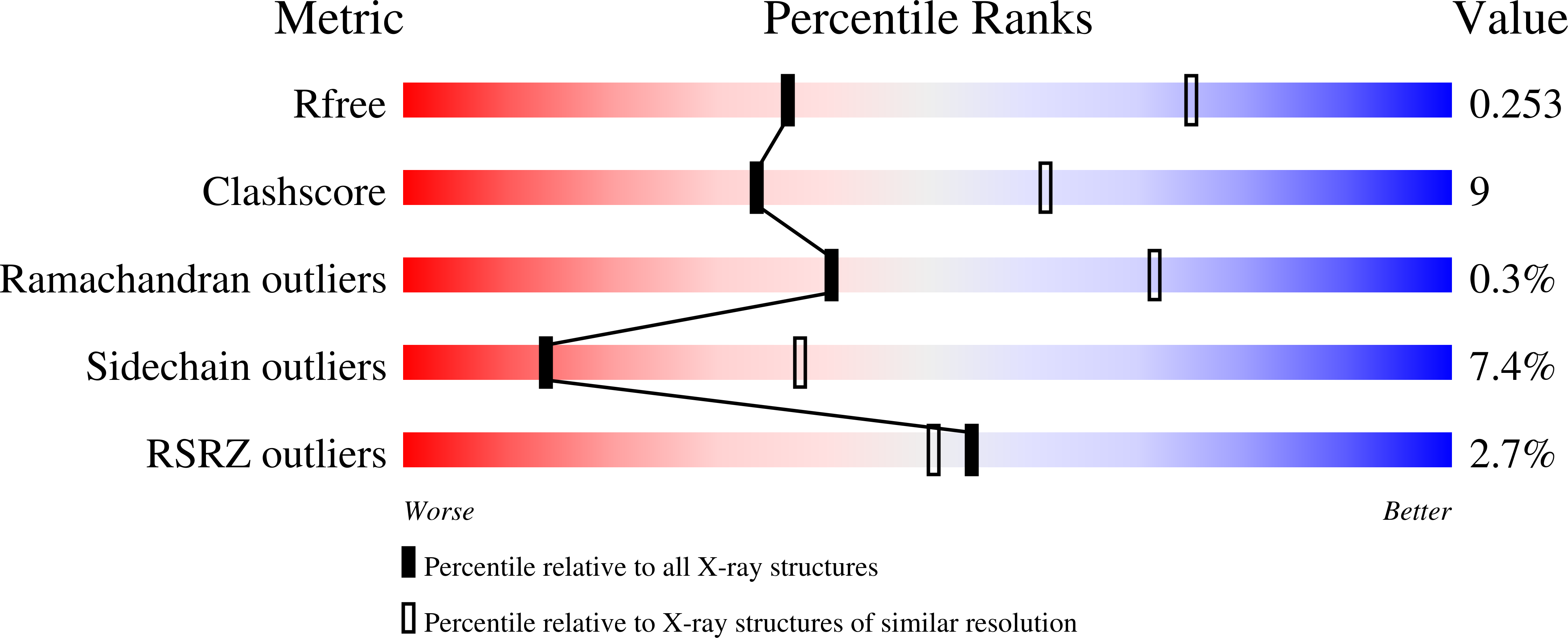
5A6C - PubMed Abstract:
Polarized epithelia form by oriented cell divisions in which the mitotic spindle aligns parallel to the epithelial plane. To orient the mitotic spindle, cortical cues trigger the recruitment of NuMA-dynein-based motors, which pull on astral microtubules via the protein LGN. We demonstrate that the junctional protein Afadin is required for spindle orientation and correct epithelial morphogenesis of Caco-2 cysts. Molecularly, Afadin binds directly and concomitantly to F-actin and to LGN. We determined the crystallographic structure of human Afadin in complex with LGN and show that it resembles the LGN-NuMA complex. In mitosis, Afadin is necessary for cortical accumulation of LGN and NuMA above the spindle poles, in an F-actin-dependent manner. Collectively, our results depict Afadin as a molecular hub governing the enrichment of LGN and NuMA at the cortex. To our knowledge, Afadin is the first-described mechanical anchor between dynein and cortical F-actin.
Organizational Affiliation:
Department of Experimental Oncology, European Institute of Oncology, Milan, Italy.