Prolyl isomerase PIN1 regulates the stability, transcriptional activity and oncogenic potential of BRD4.
Hu, X., Dong, S.H., Chen, J., Zhou, X.Z., Chen, R., Nair, S., Lu, K.P., Chen, L.F.(2017) Oncogene 36: 5177-5188
- PubMed: 28481868
- DOI: https://doi.org/10.1038/onc.2017.137
- Primary Citation of Related Structures:
5UY9 - PubMed Abstract:
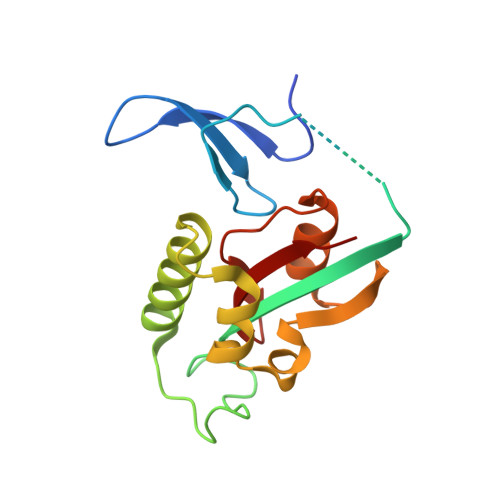
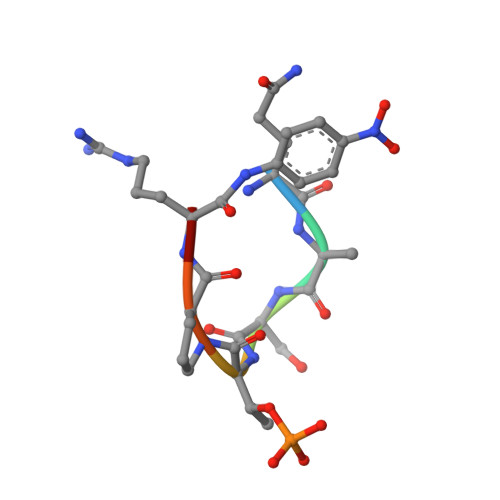
BRD4 has emerged as an important factor in tumorigenesis by promoting the transcription of genes involved in cancer development. However, how BRD4 is regulated in cancer cells remains largely unknown. Here, we report that the stability and functions of BRD4 are positively regulated by prolyl isomerase PIN1 in gastric cancer cells. PIN1 directly binds to phosphorylated threonine (T) 204 of BRD4 as revealed by peptide binding and crystallographic studies and enhances BRD4's stability by inhibiting its ubiquitination. PIN1 also catalyses the isomerization of proline 205 of BRD4 and induces its conformational change, which promotes its interaction with CDK9 and increases BRD4's transcriptional activity. Substitution of BRD4 with PIN1-binding-defective BRD4-T204A mutant in gastric cancer cells reduces BRD4's stability, attenuates BRD4-mediated gene expression by impairing its interaction with CDK9 and suppresses gastric cancer cell proliferation, migration and invasion, and tumor formation. Our results identify BRD4 as a new target of PIN1 and suggest that interfering with their interaction could be a potential therapeutic approach for cancer treatment.
Organizational Affiliation:
Department of Biochemistry, University of Illinois at Urbana-Champaign, Urbana, IL, USA.