Novel Mode of Antagonist Binding in NMDA Receptors Revealed by the Crystal Structure of the GluN1-GluN2A Ligand-Binding Domain Complexed to NVP-AAM077.
Romero-Hernandez, A., Furukawa, H.(2017) Mol Pharmacol 92: 22-29
- PubMed: 28468946
- DOI: https://doi.org/10.1124/mol.116.107912
- Primary Citation of Related Structures:
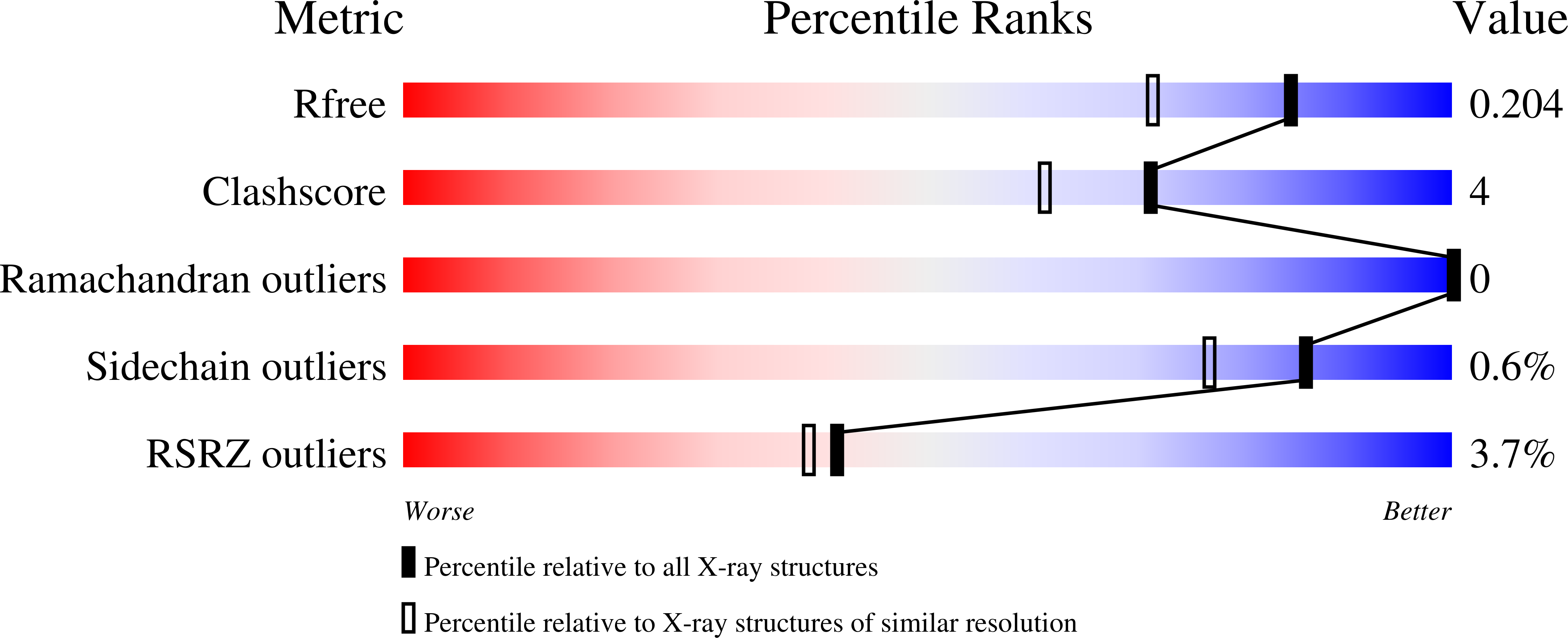
5U8C - PubMed Abstract:
Competitive antagonists against N -methyl-D-aspartate (NMDA) receptors have played critical roles throughout the history of neuropharmacology and basic neuroscience. There are currently numerous NMDA receptor antagonists containing a variety of chemical groups. Among those compounds, a GluN2-specific antagonist, (R)-[(S)-1-(4-bromo-phenyl)-ethylamino]-(2,3-dioxo-1,2,3,4-tetrahydroquinoxalin-5-yl)-methyl-phosphonic acid (NVP-AAM077), contains a unique combination of a dioxoquinoxalinyl ring, a bromophenyl group, and a phosphono group. In this study, we present the crystal structure of the isolated ligand-binding domain of the GluN1-GluN2A NMDA receptor in complex with the GluN1 agonist glycine and the GluN2A antagonist NVP-AAM077. The structure shows placement of the dioxoquinoxalinyl ring and the phosphono group of NVP-AAM077 in the glutamate-binding pocket in GluN2A and the novel interaction between the bromophenyl group and GluN1-Glu781 at the GluN1-GluN2A subunit interface. Site-directed mutagenesis of GluN1-Glu781 reduced the potency of inhibition by NVP-AAM077, thus confirming the involvement of the GluN1 subunit for binding of NVP-AAM077. The unique antagonist-binding pattern shown in this study provides a novel dimension to design and create antagonists with potential therapeutic values.
Organizational Affiliation:
Keck Structural Biology Laboratory, Cold Spring Harbor Laboratory, Cold Spring Harbor, New York (A.R.-H., H.F.); and Watson School of Biological Sciences, Cold Spring Harbor, New York (A.R.-H., H.F.).