The Antiviral Mechanism of an Influenza A Virus Nucleoprotein-Specific Single-Domain Antibody Fragment.
Hanke, L., Knockenhauer, K.E., Brewer, R.C., van Diest, E., Schmidt, F.I., Schwartz, T.U., Ploegh, H.L.(2016) mBio 7
- PubMed: 27965447
- DOI: https://doi.org/10.1128/mBio.01569-16
- Primary Citation of Related Structures:
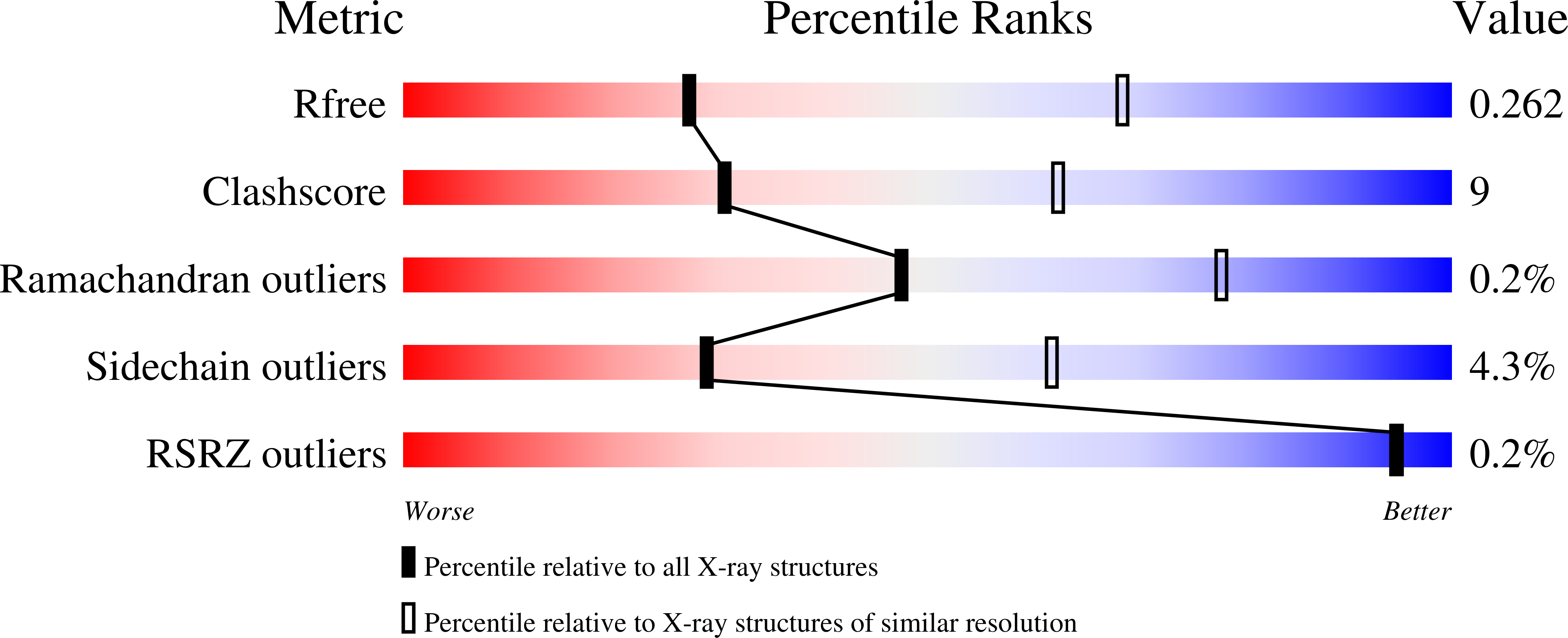
5TJW - PubMed Abstract:
Alpaca-derived single-domain antibody fragments (VHHs) that target the influenza A virus nucleoprotein (NP) can protect cells from infection when expressed in the cytosol. We found that one such VHH, αNP-VHH1, exhibits antiviral activity similar to that of Mx proteins by blocking nuclear import of incoming viral ribonucleoproteins (vRNPs) and viral transcription and replication in the nucleus. We determined a 3.2-Å crystal structure of αNP-VHH1 in complex with influenza A virus NP. The VHH binds to a nonconserved region on the body domain of NP, which has been associated with binding to host factors and serves as a determinant of host range. Several of the NP/VHH interface residues determine sensitivity of NP to antiviral Mx GTPases. The structure of the NP/αNP-VHH1 complex affords a plausible explanation for the inhibitory properties of the VHH and suggests a rationale for the antiviral properties of Mx proteins. Such knowledge can be leveraged for much-needed novel antiviral strategies. Influenza virus strains can rapidly escape from protection afforded by seasonal vaccines or acquire resistance to available drugs. Additional ways to interfere with the virus life cycle are therefore urgently needed. The influenza virus nucleoprotein is one promising target for antiviral interventions. We have previously isolated alpaca-derived single-domain antibody fragments (VHHs) that protect cells from influenza virus infection if expressed intracellularly. We show here that one such VHH exhibits antiviral activities similar to those of proteins of the cellular antiviral defense (Mx proteins). We determined the three-dimensional structure of this VHH in complex with the influenza virus nucleoprotein and identified the interaction site, which overlaps regions that determine sensitivity of the virus to Mx proteins. Our data define a new vulnerability of influenza virus, help us to better understand the cellular antiviral mechanisms, and provide a well-characterized tool to further study them.
Organizational Affiliation:
Whitehead Institute for Biomedical Research, Cambridge, Massachusetts, USA.