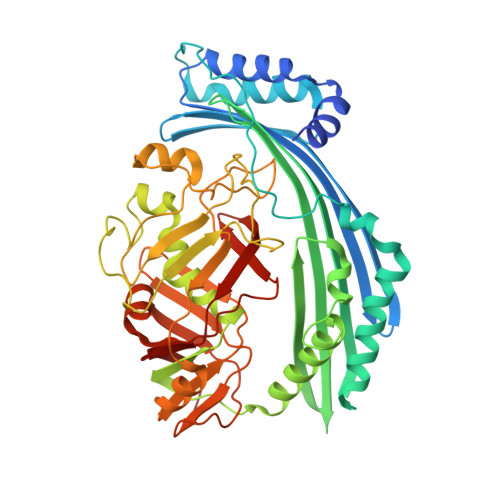
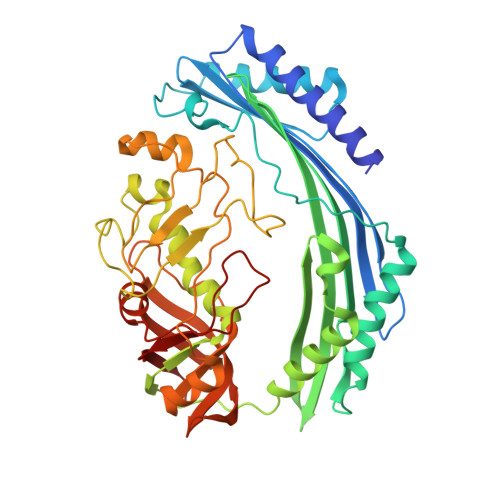
The Origins of Specificity in the Microcin-Processing Protease TldD/E.
Ghilarov, D., Serebryakova, M., Stevenson, C.E.M., Hearnshaw, S.J., Volkov, D.S., Maxwell, A., Lawson, D.M., Severinov, K.(2017) Structure 25: 1549-1561.e5
- PubMed: 28943336
- DOI: https://doi.org/10.1016/j.str.2017.08.006
- Primary Citation of Related Structures:
5NJ5, 5NJ9, 5NJA, 5NJB, 5NJC, 5NJF - PubMed Abstract:
TldD and TldE proteins are involved in the biosynthesis of microcin B17 (MccB17), an Escherichia coli thiazole/oxazole-modified peptide toxin targeting DNA gyrase. Using a combination of biochemical and crystallographic methods we show that E. coli TldD and TldE interact to form a heterodimeric metalloprotease. TldD/E cleaves the N-terminal leader sequence from the modified MccB17 precursor peptide, to yield mature antibiotic, while it has no effect on the unmodified peptide. Both proteins are essential for the activity; however, only the TldD subunit forms a novel metal-containing active site within the hollow core of the heterodimer. Peptide substrates are bound in a sequence-independent manner through β sheet interactions with TldD and are likely cleaved via a thermolysin-type mechanism. We suggest that TldD/E acts as a "molecular pencil sharpener": unfolded polypeptides are fed through a narrow channel into the active site and processively truncated through the cleavage of short peptides from the N-terminal end.
Organizational Affiliation:
Centre for Data-Intensive Biomedicine and Biotechnology, Skolkovo Institute of Science and Technology, 143026 Moscow, Russia; Institute of Gene Biology of the Russian Academy of Sciences, 119334 Moscow, Russia; Department of Biological Chemistry, John Innes Centre, Norwich Research Park, Norwich NR4 7UH, UK. Electronic address: dmitry.gilyarov@uj.edu.pl.