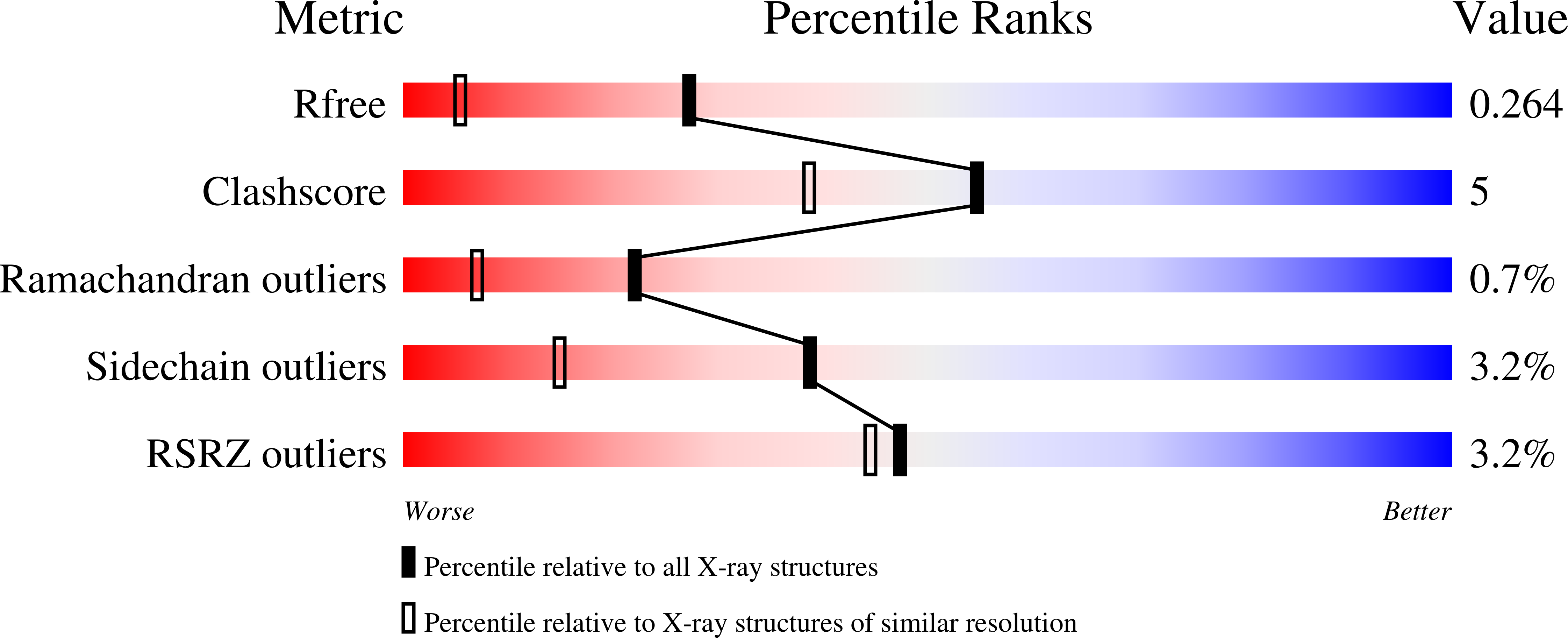
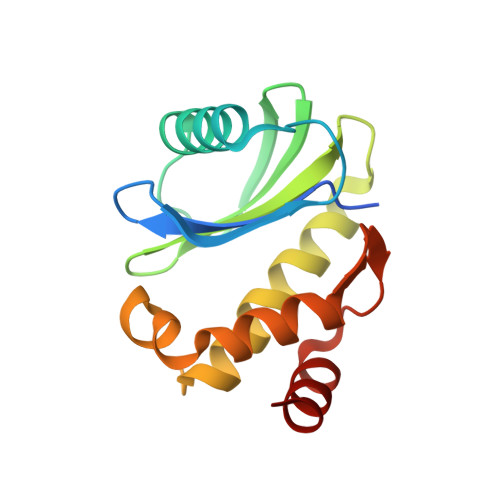
Crystal structure of truncated human coatomer protein complex subunit zeta 1 (Cop zeta 1).
Lunev, S., Semmelink, M.F., Xian, J.L., Ma, K.Y., Leenders, A.J., Domling, A.S., Shtutman, M., Groves, M.R.(2017) Acta Crystallogr F Struct Biol Commun 73: 1-8
- PubMed: 28045387
- DOI: https://doi.org/10.1107/S2053230X16018896
- Primary Citation of Related Structures:
5MC7 - PubMed Abstract:
The majority of modern anticancer approaches target DNA/protein targets involved in tumour-cell proliferation. Such approaches have a major drawback, as nonproliferating cancer cells remain unaffected and may cause relapse or remission. Human coatomer protein complex I (COPI) subunit ζ (Copζ), a component of the coat protein involved in cell apoptosis and intracellular trafficking, has recently been proposed as a potential anticancer drug target. Previous studies have shown that two different isoforms of the Copζ subunit exist in mammalian cells. While normal cells express both Copζ1 and Copζ2 isoforms, various types of tumour cells display a loss of Copζ2 expression and rely solely on Copζ1 for growth and survival. Subsequent knockdown of Copζ1 results in specific inhibition of both proliferating and dormant tumour-cell populations, with no adverse growth effects on normal cells. Therefore, a Copζ1-targeting therapy was proposed to bypass the problem of dormant cancer cells that are resistant to conventional antiproliferative drugs, which is the major cause of tumour relapse. In order to aid in structure-based inhibitor design, a crystal structure is required. In this article, the recombinant expression, purification, crystallization and crystal structure of Copζ1, as well as the expression and purification of Copζ2, are reported.
Organizational Affiliation:
Department of Drug Design, Groningen Research Institute of Pharmacy, University of Groningen, Antonius Deusinglaan 1, 9700 AD Groningen, The Netherlands.