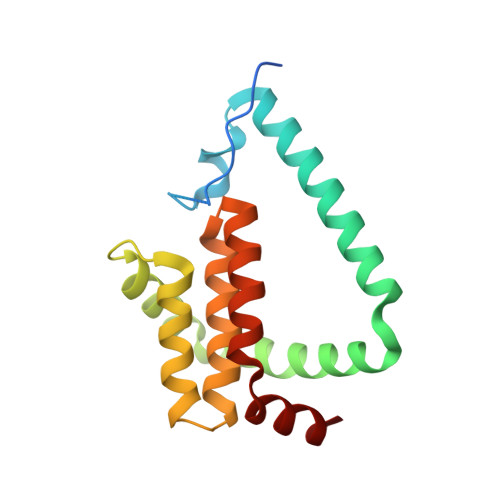
Variola Virus F1L is a Bcl-2-Like Protein that Unlike its Vaccinia Virus Counterpart Inhibits Apoptosis Independent of Bim.
Marshall, B., Puthalakath, H., Caria, S., Chugh, S., Doerflinger, M., Colman, P.M., Kvansakul, M.(2015) Cell Death Dis 6: E1680
- PubMed: 25766319
- DOI: https://doi.org/10.1038/cddis.2015.52
- Primary Citation of Related Structures:
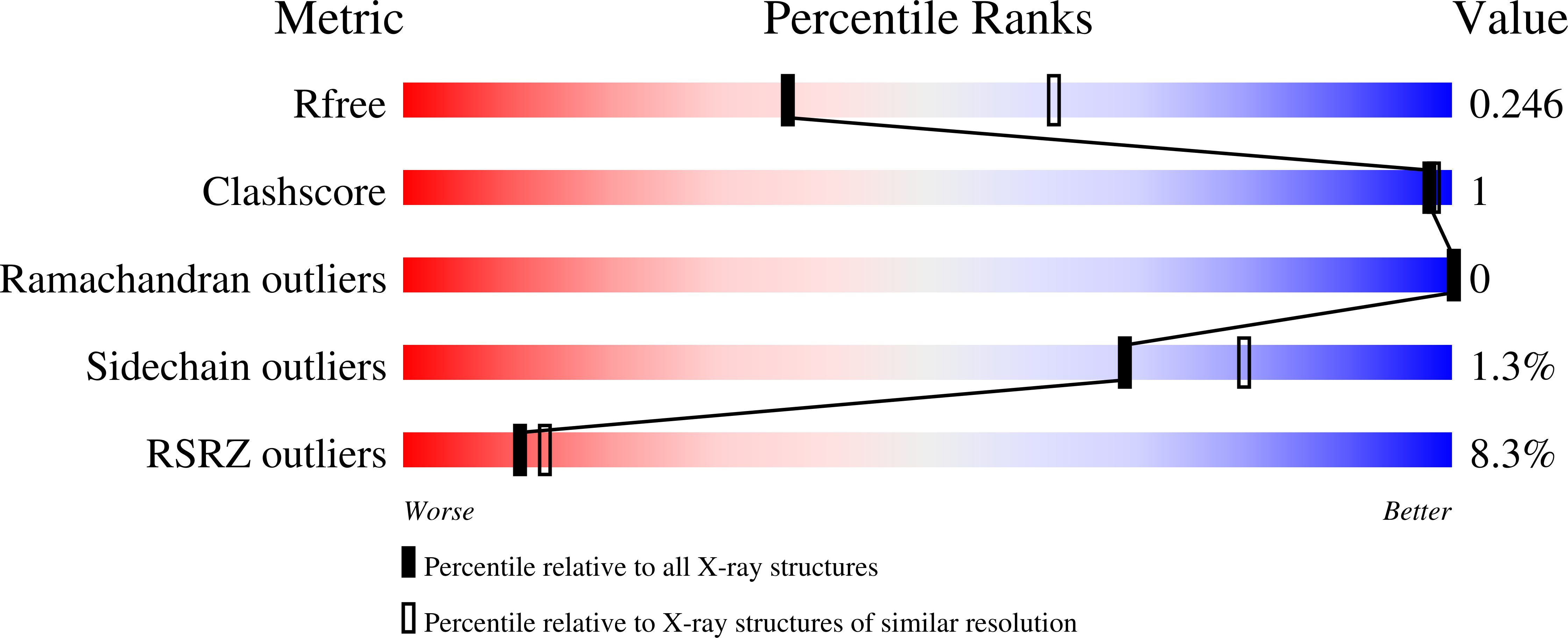
5AJJ, 5AJK - PubMed Abstract:
Subversion of host cell apoptosis is an important survival strategy for viruses to ensure their own proliferation and survival. Certain viruses express proteins homologous in sequence, structure and function to mammalian pro-survival B-cell lymphoma 2 (Bcl-2) proteins, which prevent rapid clearance of infected host cells. In vaccinia virus (VV), the virulence factor F1L was shown to be a potent inhibitor of apoptosis that functions primarily be engaging pro-apoptotic Bim. Variola virus (VAR), the causative agent of smallpox, harbors a homolog of F1L of unknown function. We show that VAR F1L is a potent inhibitor of apoptosis, and unlike all other characterized anti-apoptotic Bcl-2 family members lacks affinity for the Bim Bcl-2 homology 3 (BH3) domain. Instead, VAR F1L engages Bid BH3 as well as Bak and Bax BH3 domains. Unlike its VV homolog, variola F1L only protects against Bax-mediated apoptosis in cellular assays. Crystal structures of variola F1L bound to Bid and Bak BH3 domains reveal that variola F1L forms a domain-swapped Bcl-2 fold, which accommodates Bid and Bak BH3 in the canonical Bcl-2-binding groove, in a manner similar to VV F1L. Despite the observed conservation of structure and sequence, variola F1L inhibits apoptosis using a startlingly different mechanism compared with its VV counterpart. Our results suggest that unlike during VV infection, Bim neutralization may not be required during VAR infection. As molecular determinants for the human-specific tropism of VAR remain essentially unknown, identification of a different mechanism of action and utilization of host factors used by a VAR virulence factor compared with its VV homolog suggest that studying VAR directly may be essential to understand its unique tropism.
Organizational Affiliation:
1] Department of Biochemistry, La Trobe University, Kingsbury Drive, Melbourne, 3086 Victoria, Australia [2] La Trobe Institute for Molecular Science, La Trobe University, Kingsbury Drive, Melbourne, 3086 Victoria, Australia.