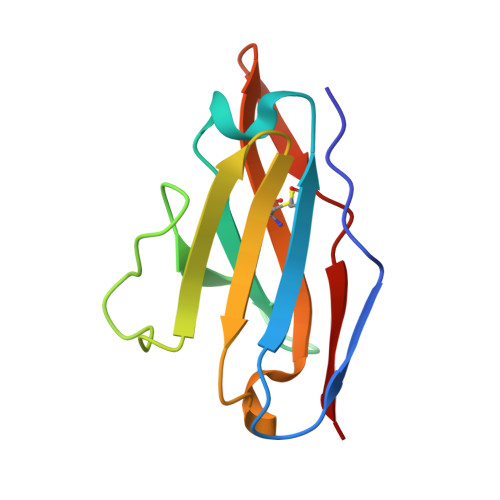
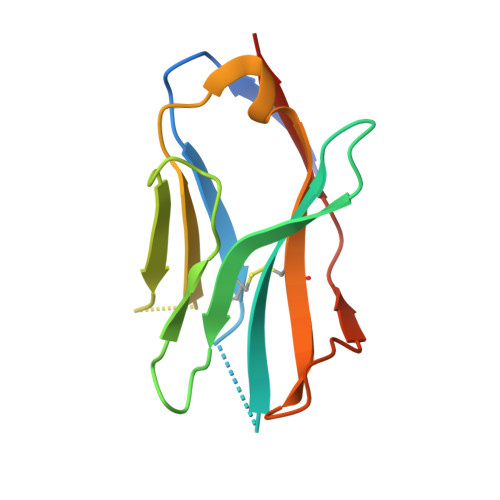
Affinity maturation of a broadly neutralizing human monoclonal antibody that prevents acute hepatitis C virus infection in mice.
Keck, Z.Y., Wang, Y., Lau, P., Lund, G., Rangarajan, S., Fauvelle, C., Liao, G.C., Holtsberg, F.W., Warfield, K.L., Aman, M.J., Pierce, B.G., Fuerst, T.R., Bailey, J.R., Baumert, T.F., Mariuzza, R.A., Kneteman, N.M., Foung, S.K.(2016) Hepatology 64: 1922-1933
- PubMed: 27641232
- DOI: https://doi.org/10.1002/hep.28850
- Primary Citation of Related Structures:
4Z0X - PubMed Abstract:
Direct-acting antivirals (DAAs) have led to a high cure rate in treated patients with chronic hepatitis C virus (HCV) infection, but this still leaves a large number of treatment failures secondary to the emergence of resistance-associated variants (RAVs). To increase the barrier to resistance, a complementary strategy is to use neutralizing human monoclonal antibodies (HMAbs) to prevent acute infection. However, earlier efforts with the selected antibodies led to RAVs in animal and clinical studies. Therefore, we identified an HMAb that is less likely to elicit RAVs for affinity maturation to increase potency and, more important, breadth of protection. Selected matured antibodies show improved affinity and neutralization against a panel of diverse HCV isolates. Structural and modeling studies reveal that the affinity-matured HMAb mediates virus neutralization, in part, by inducing conformational change to the targeted epitope, and that the maturated light chain is responsible for the improved affinity and breadth of protection. A matured HMAb protected humanized mice when challenged with an infectious HCV human serum inoculum for a prolonged period. However, a single mouse experienced breakthrough infection after 63 days when the serum HMAb concentration dropped by several logs; sequence analysis revealed no viral escape mutation.
Organizational Affiliation:
Department of Pathology, Stanford University School of Medicine, Stanford, CA.