Optimisation of a triazolopyridine based histone demethylase inhibitor yields a potent and selective KDM2A (FBXL11) inhibitor.
England, K.S., Tumber, A., Krojer, T., Scozzafava, G., Ng, S.S., Daniel, M., Szykowska, A., Che, K., von Delft, F., Burgess-Brown, N.A., Kawamura, A., Schofield, C.J., Brennan, P.E.(2014) Medchemcomm 5: 1879-1886
- PubMed: 26682034
- DOI: https://doi.org/10.1039/C4MD00291A
- Primary Citation of Related Structures:
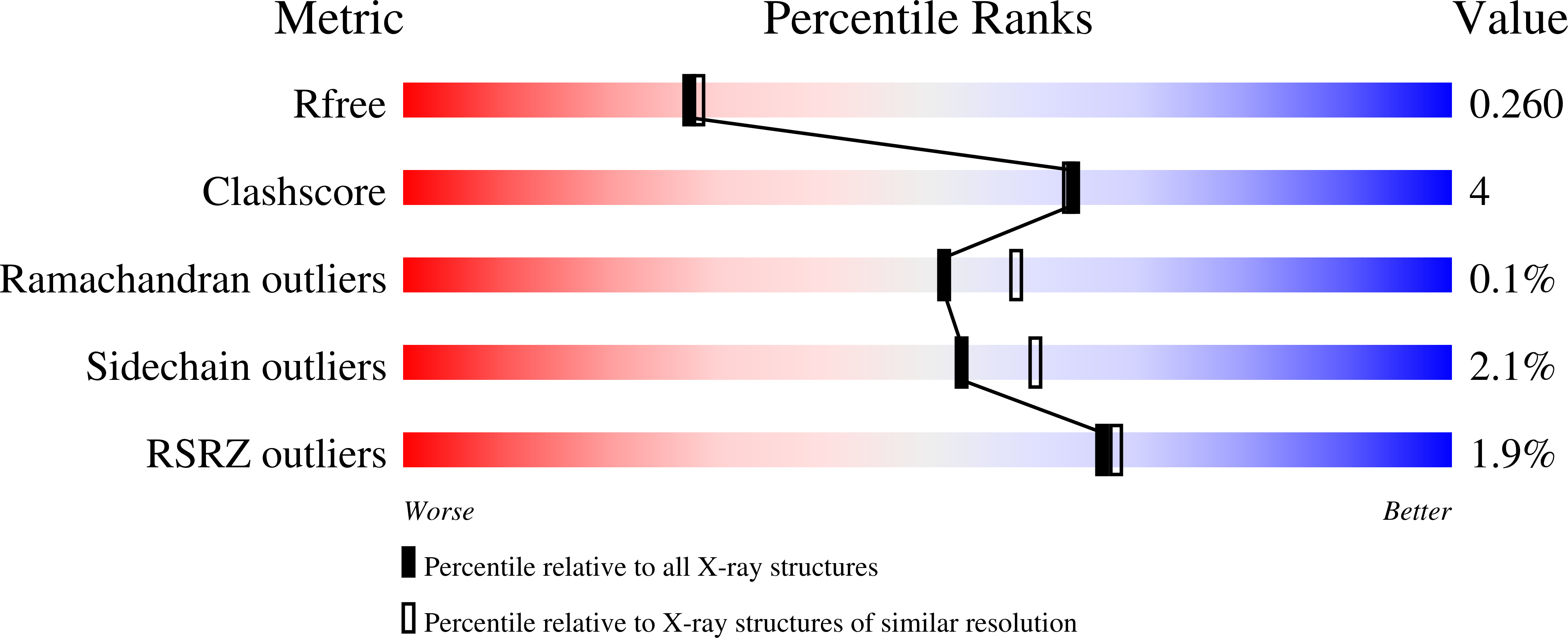
4URA - PubMed Abstract:
A potent inhibitor of the JmjC histone lysine demethylase KDM2A (compound 35 , pIC 50 7.2) with excellent selectivity over representatives from other KDM subfamilies has been developed; the discovery that a triazolopyridine compound binds to the active site of JmjC KDMs was followed by optimisation of the triazole substituent for KDM2A inhibition and selectivity.
Organizational Affiliation:
Structural Genomics Consortium, University of Oxford, Old Road Campus, Roosevelt Drive, Headington OX3 7DQ, UK.