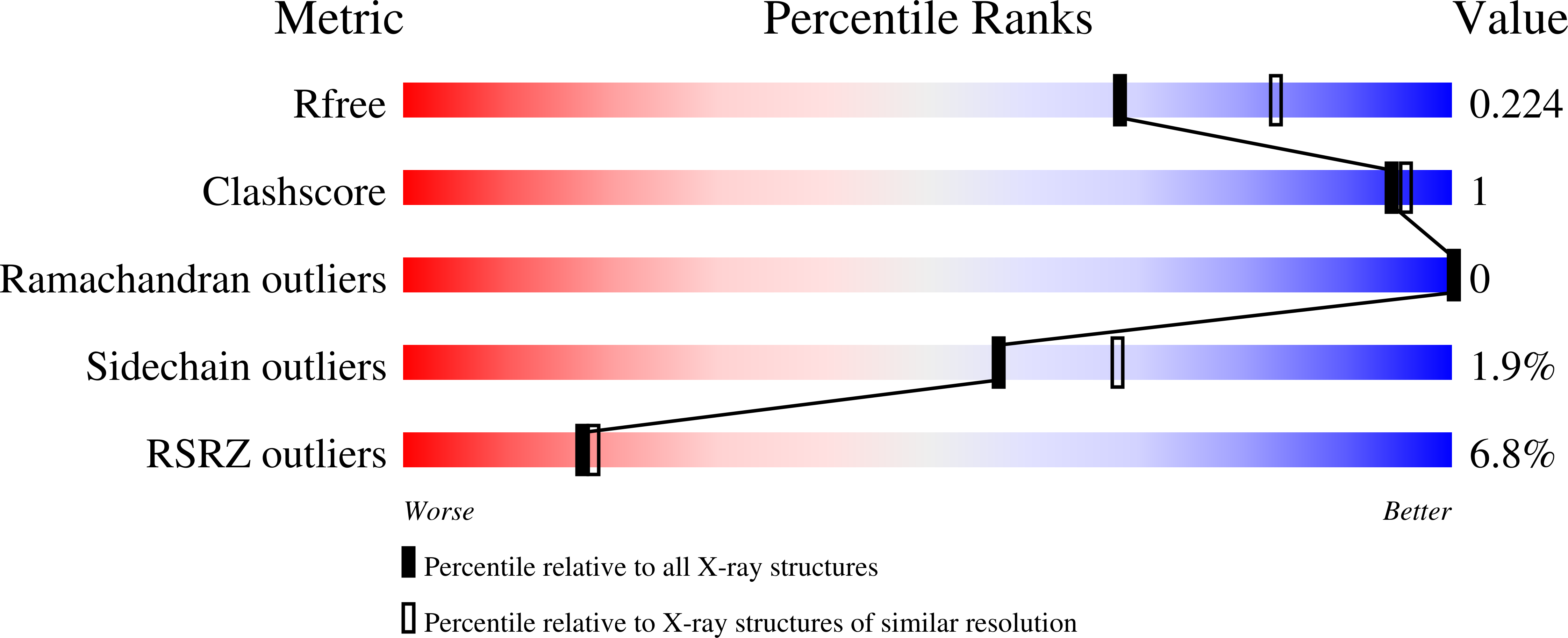
HCF-1 is cleaved in the active site of O-GlcNAc transferase.
Lazarus, M.B., Jiang, J., Kapuria, V., Bhuiyan, T., Janetzko, J., Zandberg, W.F., Vocadlo, D.J., Herr, W., Walker, S.(2013) Science 342: 1235-1239
- PubMed: 24311690
- DOI: https://doi.org/10.1126/science.1243990
- Primary Citation of Related Structures:
4N39, 4N3A, 4N3B, 4N3C - PubMed Abstract:
Host cell factor-1 (HCF-1), a transcriptional co-regulator of human cell-cycle progression, undergoes proteolytic maturation in which any of six repeated sequences is cleaved by the nutrient-responsive glycosyltransferase, O-linked N-acetylglucosamine (O-GlcNAc) transferase (OGT). We report that the tetratricopeptide-repeat domain of O-GlcNAc transferase binds the carboxyl-terminal portion of an HCF-1 proteolytic repeat such that the cleavage region lies in the glycosyltransferase active site above uridine diphosphate-GlcNAc. The conformation is similar to that of a glycosylation-competent peptide substrate. Cleavage occurs between cysteine and glutamate residues and results in a pyroglutamate product. Conversion of the cleavage site glutamate into serine converts an HCF-1 proteolytic repeat into a glycosylation substrate. Thus, protein glycosylation and HCF-1 cleavage occur in the same active site.
Organizational Affiliation:
Department of Microbiology and Immunobiology, Harvard Medical School, Boston, MA 02115, USA.