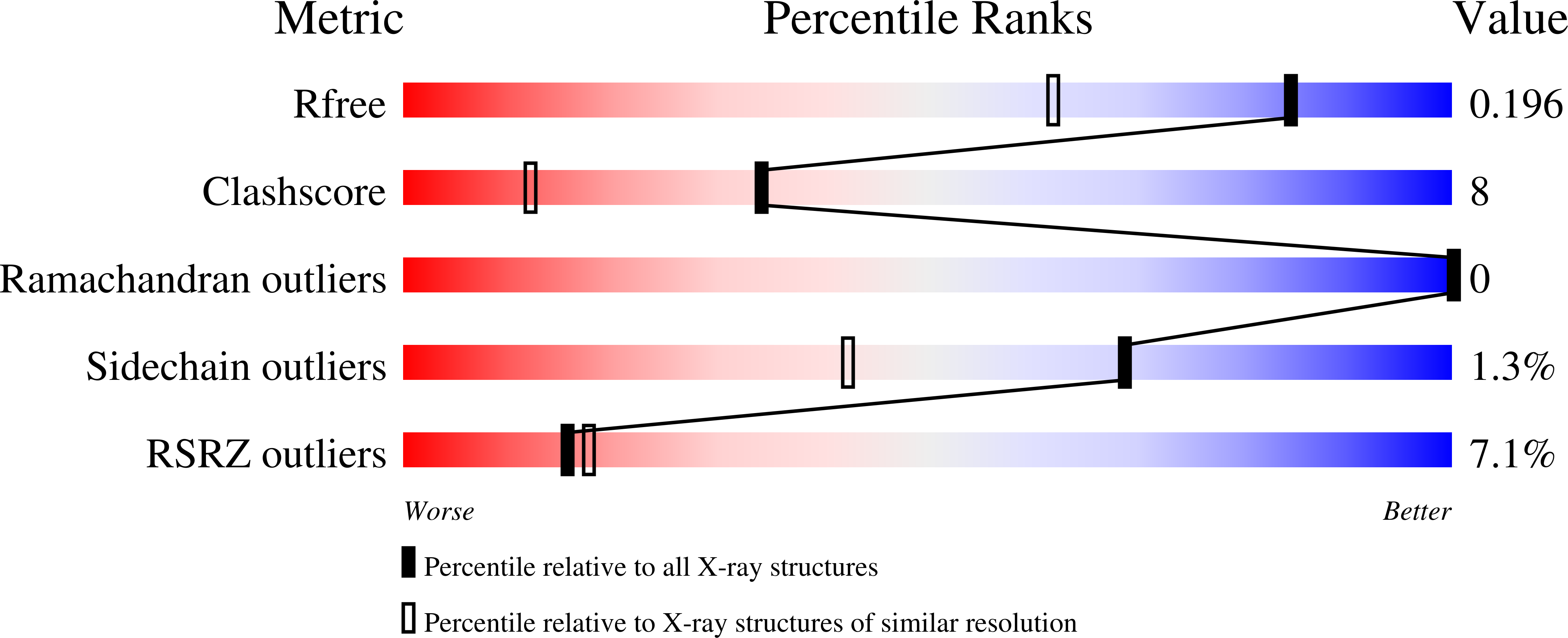
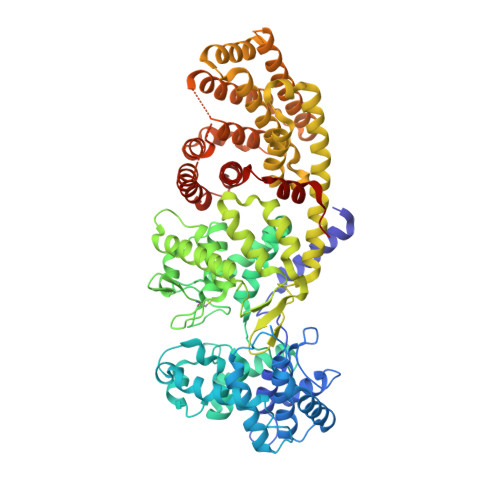
1.55 angstrom -resolution structure of ent-copalyl diphosphate synthase and exploration of general acid function by site-directed mutagenesis.
Koksal, M., Potter, K., Peters, R.J., Christianson, D.W.(2013) Biochim Biophys Acta 1840: 184-190
- PubMed: 24036329
- DOI: https://doi.org/10.1016/j.bbagen.2013.09.004
- Primary Citation of Related Structures:
4LIX - PubMed Abstract:
The diterpene cyclase ent-copalyl diphosphate synthase (CPS) catalyzes the first committed step in the biosynthesis of gibberellins. The previously reported 2.25Å resolution crystal structure of CPS complexed with (S)-15-aza-14,15-dihydrogeranylgeranyl thiolodiphosphate (1) established the αβγ domain architecture, but ambiguities regarding substrate analog binding remained.
Organizational Affiliation:
Roy and Diana Vagelos Laboratories, Department of Chemistry, University of Pennsylvania, Philadelphia, PA 19104-6323, USA.